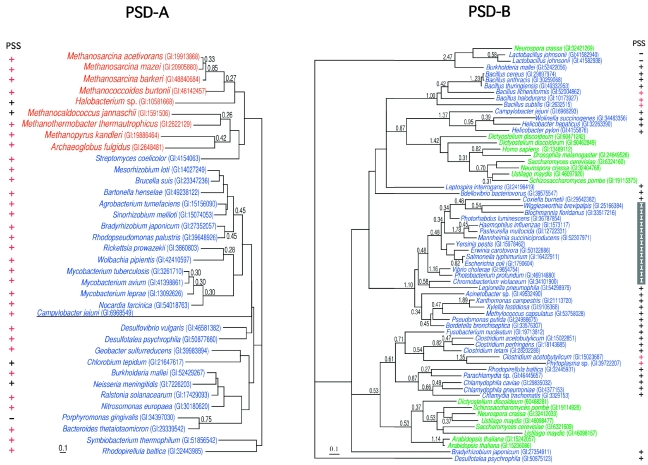
Figure 4.
Two unrooted maximum likelihood trees of phosphatidylserine synthase/archaetidylserine synthase (PSS/ASS). Each sequence is indicated by the source name and the GI number from the National Center for Biotechnology Information. The colors for each species are the same as in Figure 3. The reliability index of a cluster is only shown when the value is equal to or greater than 0.25. The log likelihoods were –7157.86 and –18485.60 for phosphatidylserine decarboxylase-A and -B (PSD-A or PSD-B), respectively. The optimal shape parameter alpha was 0.85 (145 sites) for PSD-A and 1.13 (197 sites) for PSD-B. The presence or absence of ASS or PSS-II in each species is indicated by a symbol, ‘+’ or ‘–’, near the source name. The symbol, ‘+’, colored red, shows that the genes or open reading frames for PSS/ASS and PSD/ASD are tandemly encoded. When the species uses PSS-I instead of PSS-II, the symbol ‘+’ is replaced with ‘I’.