Abstract
With rapidly growing interest in the urine proteome, methods for reducing sample complexity are becoming increasingly important. Depletion strategies for removal of high-abundance proteins from human urine have not been reported. A commercial kit designed for depletion of abundant proteins from plasma was evaluated for removing top proteins from urine of patients with proteinuria. The number of low-abundance proteins identified in urine after depletion increased nearly 2.5-fold.
Keywords: affinity chromatography, protein depletion, proteinuria, proteomics, urine
INTRODUCTION
The detection and identification of trace amounts of proteins in complex samples is a major challenge in bio-markers discovery and validation. Depletion of proteins present in high concentrations is one common approach for improved detection of low-abundance proteins, which may serve as potential biomarkers.1–3 The depletion strategy often employs antibody affinity chemistry using commercially available kits.4,5
Samples of interest for detection of protein biomarkers are typically serum or plasma. Thus, commercial kits for the depletion of abundant proteins are generally developed and optimized for plasma or serum samples. However, with the rapidly growing interest in the human urine proteome, methods for reducing the complexity of urine samples are needed.6–8 To date, depletion strategies for removal of high-abundance proteins from human urine have not been reported.
Fractionation of samples has been recognized as one common approach for reducing complexity and making the samples amenable for instrumental analysis. One of the most efficient methods for reduction of sample complexity is affinity capture of abundant proteins followed by analysis of the remaining fraction.1,4,9
Because of the simplicity and noninvasiveness of collection, urine is an attractive sample type for many diagnostic tests. Furthermore, urine contains a relatively small number of proteins typically present at low concentrations, and thus a simpler matrix for detecting proteins as compared to serum or plasma. However, in some human diseases excess protein is found in the urine, as can occur in patients with compromised kidney function. As a result, many of the proteins normally present in blood will also be excreted into the urine. This condition, known as proteinuria, is often observed in acute inflammation, acute urinary tract infection, amyloidosis, diabetic nephropathy, kidney failure, multiple myeloma, nephrotic syndrome, and severe yeast infections. 10–14
When searching for diagnostic markers of the above diseases and conditions, plasma proteins present at high concentrations in urine samples can hinder the ability to detect potential protein biomarkers present at low concentrations. Depletion of high-abundance plasma proteins from urine is one approach that would simplify sample complexity and improve the chances of finding potential biomarkers. In this study we report the use of a commercially manufactured protein depletion kit for the removal of the six most abundant human plasma proteins from urine samples.
EXPERIMENTAL PROCEDURES
To evaluate the commercial protein depletion kit, human serum samples were prepared and processed according to the manufacturer’s instructions. Test samples of pooled human urine were then prepared. A protein-containing urine pool was collected from the urine of patients with proteinuria. For the negative control, pooled urine samples from healthy individuals were used. All samples were de-identified in accordance with a University of Utah Institutional Review Board approved protocol.
The multiple affinity removal (MARS) column for the depletion of 6 high-abundant proteins (Agilent Technologies, Santa Clara, CA, Part #5185–5984) contains affinity binders for the depletion of albumin, transferrin, haptoglobin, IgG, IgA, and alpha-1 antitrypsin. Samples were prepared by ultrafiltration (Amicon Ultra-4 filters, 5 kDa cutoff, Millipore, Billerica, MA), then processed with the recommended column run cycle consisting of loading the sample (serum or urine), collecting the flow-through (depleted fraction) proteins, washing, and eluting bound proteins.
The efficiency of the depletion of the proteins from urine was confirmed using SDS-PAGE. Where possible, equal loading of total protein onto the depletion column and for gel analysis was employed. After electrophoresis, the gel was silver stained (Invitrogen, Carlsbad, CA) and imaged.
Samples were processed by alkylating cysteine residues with iodoacetamide, then either in-gel or in-solution tryptic digest for 18 h at 37°C. The samples were then analyzed on the Agilent 6510 Q-TOF (Agilent Technologies, Santa Clara, CA) equipped with a ChipCube (G2240A) and an Agilent 1200 nano-HPLC system using a 30-min acetonitrile gradient (5%–40%) and C18 reversed-phase separation. Data acquisition (2 GHz extended dynamic range) was performed with MassHunter Q-TOF Acquisition software B.01.03 with an acquisition rate of 3 scans/s followed by tandem mass spectrometry (MS/MS) scans of the three most intense ions using the acquisition rate of 2 scans/s. Exclusion was set for 12 s after two consecutive MS/MS scans of a precursor ion. The time-of-flight (TOF) analyzer was tuned to a resolution of 12,000 and calibrated prior to each experiment for a mass accuracy < 2 ppm.
The acquired MS/MS spectra were searched using Spectrum Mill MS Proteomics Workbench Rev A.03.03.075 (Agilent) against the SwissProt human database (v 13.2–53,541 entries). Mass tolerance was set to 20 ppm for precursor ions and 50 ppm for fragment ions. An enzyme-specific search (trypsin) was used with two missed cleavages allowed. Variable carbamidomethylation (Da 57.0214) was also included for peptide scoring during the database search. Search results were filtered to approximately 5% error using Spectrum Mill’s autovalidation tool, with an individual peptide score threshold of 10 or summed score of 15.
RESULTS
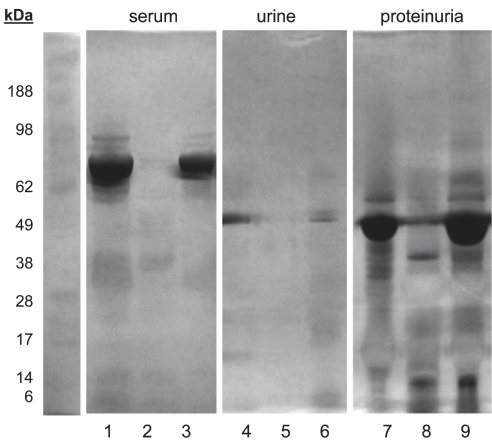
High-abundance proteins were removed from human serum using the Agilent MARS column. The depletion was performed according to the method recommended by the manufacturer for serum samples. Figure 1 shows the silver-stained SDS gel of the control serum sample (lane 1), flow-through of the depletion column (lane 2), and the eluted fraction of serum proteins retained by the column (lane 3).
FIGURE 1.
Depletion of high-abundance proteins from serum (lanes 1–3) and urine (lanes 4–9) samples. Lane 1: Serum control; lane 2: depletion column flow-through; lane 3: column elution of the bound serum proteins. Lane 4: normal urine control; lane 5: column flow-through; lane 6: column elution. Lane 7: proteinuria control; lane 8: flow-through fraction; lane 9: column elution of the bound protein fraction.
Performance of the depletion kit was also compared between serum and urine samples. The pool of urine samples collected from healthy individuals contained traces of proteins (lane 4), which were removed in the flow-through fraction (lane 5) and accounted for in the eluted fraction (lane 6). The gel analysis of proteinuria samples contained large amounts of proteins (lane 7), which were removed in the flow-through fraction (lane 8) and seen in the eluted fraction (lane 9).
To further evaluate the performance of depletion in urine, the total number of proteins identified by liquid chromatography (LC)-MS/MS analysis of urine samples was compared. While only 29 proteins were identified in urine from healthy individuals (Fig. 1, lane 4 and Table 1), some 60 proteins were identified in urine from patients with proteinuria (Fig. 1, lane 7 and Table 2). However, after depletion of high-abundance proteins, 142 proteins were identified (Fig. 1, lane 8 and Table 3). Table 4 summarizes these findings.
TABLE 1.
Human Proteins Identified in Urine of Healthy Individuals
| Group No. | No. Spectra | No. Unique PEPs | % Coverage | Unique Score | Accession | Protein |
|---|---|---|---|---|---|---|
| 1 | 20 | 10 | 20 | 147.9 | P02768 | Serum albumin precursor |
| 2 | 18 | 7 | 65 | 117.5 | P62988 | Ubiquitin |
| 3 | 4 | 3 | 2 | 41.8 | P02452 | Collagen alpha-1(I) chain precursor |
| 4 | 3 | 2 | 8 | 38.7 | P10645 | Chromogranin-A precursor |
| 5 | 3 | 3 | 12 | 29.9 | P11684 | Uteroglobin precursor |
| 6 | 4 | 3 | 12 | 29.6 | P01009 | Alpha-1-antitrypsin precursor |
| 7 | 3 | 2 | 4 | 28.5 | P04080 | Cystatin-B |
| 8 | 13 | 4 | 33 | 27.2 | P01834 | Ig kappa chain C region |
| 9 | 2 | 2 | 31 | 21.6 | Q9UGM3 | Deleted in malignant brain tumors 1 protein precursor |
| 10 | 5 | 2 | 1 | 21.4 | P01344 | Insulin-like growth factor II precursor |
| 11 | 2 | 2 | 4 | 21.2 | P02814 | Submaxillary gland androgen-regulated protein 3 homolog B |
| 12 | 7 | 2 | 39 | 20.7 | P68133 | Actin, alpha skeletal muscle |
| 13 | 2 | 3 | 2 | 20.4 | P68363 | Tubulin alpha-1B chain |
| 14 | 2 | 2 | 3 | 20.3 | P36578 | 60S ribosomal protein L4 |
| 15 | 2 | 2 | 4 | 19.7 | P18135 | Ig kappa chain V-III region HAH precursor |
| 16 | 2 | 2 | 6 | 19.7 | P02750 | Leucine-rich alpha-2-glycoprotein precursor |
| 17 | 2 | 2 | 2 | 19.0 | P02760 | AMBP protein precursor (contains alpha-1-microglobulin) |
| 18 | 2 | 2 | 4 | 18.9 | P00738 | Haptoglobin precursor (contains haptoglobin alpha chain) |
| 19 | 2 | 2 | 1 | 18.9 | P01602 | Ig kappa chain V-I region HK102 precursor |
| 20 | 2 | 2 | 13 | 18.4 | P10153 | Nonsecretory ribonuclease precursor |
| 21 | 2 | 2 | 9 | 18.2 | P01842 | Ig lambda chain C regions |
| 22 | 2 | 2 | 27 | 17.0 | P02753 | Plasma retinol-binding protein precursor |
| 23 | 2 | 2 | 4 | 16.8 | P07602 | Proactivator polypeptide precursor (contains Saposin-A) |
| 24 | 2 | 2 | 2 | 16.8 | P01703 | Ig lambda chain V-I region NEWM |
| 25 | 2 | 2 | 10 | 16.1 | Q12907 | Vesicular integral-membrane protein VIP36 precursor |
| 26 | 4 | 2 | 2 | 15.3 | P01857 | Ig gamma-1 chain C region |
| 27 | 3 | 2 | 3 | 15.1 | P19971 | Thymidine phosphorylase precursor |
| 28 | 2 | 2 | 2 | 15.0 | Q99459 | Cell division cycle 5-like protein |
| 29 | 3 | 2 | 2 | 15.0 | P80748 | Ig lambda chain V-III region LOI |
TABLE 2.
Human Proteins Identified in Urine of Individuals with Proteinuria
| Group No. | No. Spectra | No. Unique PEPs | % Coverage | Unique Score | Accession | Protein |
|---|---|---|---|---|---|---|
| 1 | 42 | 12 | 17 | 218.0 | P02768 | Serum albumin precursor |
| 2 | 28 | 6 | 9 | 107.8 | P07911 | Uromodulin precursor |
| 3 | 11 | 5 | 1 | 98.0 | P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein |
| 4 | 11 | 4 | 4 | 75.4 | P01133 | Pro-epidermal growth factor precursor |
| 5 | 7 | 3 | 7 | 58.9 | P19961 | Alpha-amylase 2B precursor |
| 6 | 6 | 3 | 8 | 56.5 | P10909 | Clusterin precursor |
| 7 | 6 | 3 | 5 | 56.3 | Q6EMK4 | Vasorin precursor |
| 8 | 7 | 3 | 5 | 52.5 | P01042 | Kininogen-1 precursor |
| 9 | 15 | 4 | 16 | 52.3 | P05090 | Apolipoprotein D precursor |
| 10 | 4 | 2 | 3 | 40.3 | P55290 | Cadherin-13 precursor |
| 11 | 6 | 2 | 5 | 37.6 | P01876 | Ig alpha-1 chain C region |
| 12 | 7 | 2 | 9 | 37.0 | P10451 | Osteopontin precursor |
| 13 | 6 | 2 | 3 | 36.1 | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 precursor |
| 14 | 6 | 2 | 3 | 34.0 | Q08380 | Galectin-3-binding protein precursor |
| 15 | 4 | 2 | 3 | 33.4 | P01833 | Polymeric-immunoglobulin receptor precursor |
| 16 | 3 | 2 | 4 | 32.0 | P68871 | Hemoglobin subunit beta |
| 17 | 2 | 2 | 9 | 27.2 | P01009 | Alpha-1-antitrypsin precursor |
| 18 | 4 | 2 | 4 | 25.2 | P10153 | Nonsecretory ribonuclease precursor |
| 19 | 2 | 2 | 18 | 25.1 | P07288 | Prostate-specific antigen precursor |
| 20 | 3 | 3 | 8 | 24.9 | P01834 | Ig kappa chain C region |
| 21 | 2 | 2 | 1 | 24.6 | Q7Z5L0 | Vitelline membrane outer layer protein 1 homolog precursor |
| 22 | 2 | 2 | 2 | 24.4 | P10253 | Lysosomal alpha-glucosidase precursor |
| 23 | 5 | 2 | 1 | 24.0 | Q9HCU0 | Endosialin precursor |
| 24 | 4 | 3 | 5 | 23.7 | P12109 | Collagen alpha-1(VI) chain precursor |
| 25 | 4 | 2 | 8 | 22.9 | P05452 | Tetranectin precursor |
| 26 | 6 | 2 | 3 | 22.5 | P41222 | Prostaglandin-H2 D-isomerase precursor |
| 27 | 2 | 2 | 4 | 21.2 | P04004 | Vitronectin precursor |
| 28 | 2 | 2 | 4 | 20.7 | Q6GTX8 | Leukocyte-associated immunoglobulin-like receptor 1 precursor |
| 29 | 2 | 2 | 2 | 20.6 | P02774 | Vitamin D-binding protein precursor |
| 30 | 4 | 3 | 9 | 20.6 | P01859 | Ig gamma-2 chain C region |
| 31 | 3 | 3 | 2 | 20.5 | P06310 | Ig kappa chain V-II region RPMI 6410 precursor |
| 32 | 2 | 2 | 3 | 20.2 | O94919 | Endonuclease domain-containing 1 protein precursor |
| 33 | 4 | 2 | 4 | 20.0 | P15309 | Prostatic acid phosphatase precursor |
| 34 | 2 | 2 | 4 | 20.0 | P06870 | Kallikrein-1 precursor |
| 35 | 2 | 2 | 15 | 19.9 | O75594 | Peptidoglycan recognition protein precursor |
| 36 | 2 | 2 | 1 | 19.4 | P01766 | Ig heavy chain V-III region BRO |
| 37 | 4 | 2 | 2 | 19.2 | P04264 | Keratin, type II cytoskeletal 1 |
| 38 | 2 | 2 | 7 | 19.2 | Q12907 | Vesicular integral-membrane protein VIP36 precursor |
| 39 | 2 | 2 | 2 | 19.0 | P01781 | Ig heavy chain V-III region GAL |
| 40 | 4 | 2 | 2 | 19.0 | O00187 | Mannan-binding lectin serine protease 2 precursor |
| 41 | 3 | 3 | 1 | 18.8 | P01871 | Ig mu chain C region |
| 42 | 2 | 2 | 5 | 18.8 | P16070 | CD44 antigen precursor |
| 43 | 4 | 2 | 2 | 18.5 | P25311 | Zinc-alpha-2-glycoprotein precursor |
| 44 | 2 | 2 | 1 | 18.4 | P05155 | Plasma protease C1 inhibitor precursor |
| 45 | 2 | 2 | 2 | 17.4 | P14543 | Nidogen-1 precursor |
| 46 | 6 | 2 | 1 | 17.0 | Q9GZM5 | Protein YIPF3 |
| 47 | 3 | 2 | 2 | 16.9 | P35908 | Keratin, type II cytoskeletal 2 epidermal |
| 48 | 2 | 2 | 5 | 16.9 | P08571 | Monocyte differentiation antigen CD14 precursor |
| 49 | 2 | 2 | 1 | 16.8 | P18827 | Syndecan-1 precursor |
| 50 | 2 | 2 | 2 | 16.6 | Q6UVK1 | Chondroitin sulfate proteoglycan 4 precursor |
| 51 | 3 | 3 | 1 | 16.6 | P69905 | Hemoglobin subunit alpha |
| 52 | 6 | 2 | 1 | 16.3 | P02760 | AMBP protein precursor [Contains: Alpha-1-microglobulin] |
| 53 | 4 | 2 | 1 | 16.1 | P06396 | Gelsolin precursor |
| 54 | 2 | 2 | 1 | 15.1 | Q6XZF7 | Dynamin-binding protein |
| 55 | 3 | 3 | 1 | 15.1 | P02751 | Fibronectin precursor |
| 56 | 2 | 2 | 2 | 15.0 | Q00796 | Sorbitol dehydrogenase |
| 57 | 2 | 2 | 3 | 15.0 | P12830 | Epithelial-cadherin precursor |
| 58 | 3 | 2 | 9 | 15.0 | P02750 | Leucine-rich alpha-2-glycoprotein precursor |
| 59 | 2 | 2 | 2 | 15.0 | O75144 | ICOS ligand precursor |
| 60 | 6 | 2 | 1 | 15.0 | Q8IZQ5 | Selenoprotein H |
TABLE 3.
Human Proteins Identified in Urine of Individuals with Proteinuria Using a Depletion Strategy
| Group No. | No. Spectra | No. Unique PEPs | % Coverage | Unique Score | Accession | Protein |
|---|---|---|---|---|---|---|
| 1 | 80 | 14 | 46 | 261.1 | P25311 | Zinc-alpha-2-glycoprotein precursor |
| 2 | 33 | 10 | 11 | 161.7 | P15144 | Aminopeptidase N |
| 3 | 80 | 8 | 69 | 152.5 | P62988 | Ubiquitin |
| 4 | 66 | 6 | 27 | 117.2 | P02763 | Alpha-1-acid glycoprotein 1 precursor |
| 5 | 47 | 6 | 30 | 114.1 | P10451 | Osteopontin precursor |
| 6 | 92 | 7 | 21 | 107.4 | P02760 | AMBP protein precursor (contains alpha-1-microglobulin) |
| 7 | 30 | 6 | 18 | 105.7 | P80188 | Neutrophil gelatinase-associated lipocalin precursor |
| 8 | 17 | 6 | 43 | 103.3 | P27487 | Dipeptidyl peptidase 4 |
| 9 | 13 | 7 | 7 | 102.0 | P10253 | Lysosomal alpha-glucosidase precursor |
| 10 | 23 | 6 | 10 | 100.8 | P08571 | Monocyte differentiation antigen CD14 precursor |
| 11 | 21 | 5 | 22 | 96.7 | P15586 | N-acetylglucosamine-6-sulfatase precursor |
| 12 | 42 | 5 | 9 | 89.4 | P07602 | Proactivator polypeptide precursor (contains saposin-A) |
| 13 | 12 | 6 | 8 | 88.3 | O43451 | Maltase-glucoamylase, intestinal (includes maltase) |
| 14 | 32 | 6 | 9 | 83.3 | P07911 | Uromodulin precursor |
| 15 | 15 | 5 | 3 | 81.7 | P07339 | Cathepsin D precursor |
| 16 | 13 | 5 | 8 | 81.3 | P01011 | Alpha-1-antichymotrypsin precursor |
| 17 | 14 | 5 | 15 | 78.6 | P04746 | Pancreatic alpha-amylase precursor |
| 18 | 9 | 4 | 15 | 77.4 | Q13228 | Selenium-binding protein 1 |
| 19 | 15 | 5 | 10 | 75.1 | P01833 | Polymeric-immunoglobulin receptor precursor |
| 20 | 23 | 4 | 11 | 74.7 | P02768 | Serum albumin precursor |
| 21 | 20 | 4 | 7 | 73.6 | P17900 | Ganglioside GM2 activator precursor |
| 22 | 13 | 5 | 23 | 70.7 | P00915 | Carbonic anhydrase 1 |
| 23 | 9 | 5 | 26 | 68.9 | Q92820 | Gamma-glutamyl hydrolase precursor |
| 24 | 7 | 3 | 21 | 63.5 | P02452 | Collagen alpha-1(I) chain precursor |
| 25 | 7 | 4 | 2 | 61.2 | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 precursor |
| 26 | 7 | 3 | 6 | 60.9 | P22352 | Glutathione peroxidase 3 precursor |
| 27 | 10 | 3 | 19 | 59.1 | P07686 | Beta-hexosaminidase beta chain precursor |
| 28 | 20 | 3 | 5 | 57.4 | P05451 | Lithostathine 1 alpha precursor |
| 29 | 18 | 2 | 18 | 54.2 | P01593 | Ig kappa chain V-I region AG |
| 30 | 10 | 3 | 30 | 53.3 | Q9Y5Y7 | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor |
| 31 | 7 | 3 | 8 | 51.8 | P09603 | Macrophage colony-stimulating factor 1 precursor |
| 32 | 12 | 3 | 7 | 51.3 | O00584 | Ribonuclease T2 precursor |
| 33 | 6 | 3 | 10 | 51.2 | P00751 | Complement factor B precursor |
| 34 | 13 | 3 | 6 | 50.9 | P01620 | Ig kappa chain V-III region SIE |
| 35 | 25 | 3 | 38 | 50.7 | P41222 | Prostaglandin-H2 D-isomerase precursor |
| 36 | 13 | 3 | 20 | 50.3 | Q08380 | Galectin-3-binding protein precursor |
| 37 | 15 | 2 | 5 | 48.2 | P01834 | Ig kappa chain C region |
| 38 | 8 | 3 | 34 | 47.9 | P12830 | Epithelial-cadherin precursor |
| 39 | 12 | 3 | 5 | 45.9 | P13473 | Lysosome-associated membrane glycoprotein 2 precursor |
| 40 | 9 | 3 | 7 | 44.8 | P05155 | Plasma protease C1 inhibitor precursor |
| 41 | 7 | 2 | 5 | 44.0 | P07478 | Trypsin-2 precursor |
| 42 | 6 | 2 | 7 | 43.1 | Q13231 | Chitotriosidase-1 precursor |
| 43 | 8 | 2 | 5 | 43.0 | Q9h299 | SH3 domain-binding glutamic acid-rich-like protein 3 |
| 44 | 6 | 3 | 20 | 42.3 | P06865 | Beta-hexosaminidase alpha chain precursor |
| 45 | 11 | 3 | 5 | 42.2 | P02753 | Plasma retinol-binding protein precursor |
| 46 | 4 | 2 | 24 | 40.9 | O94919 | Endonuclease domain-containing 1 protein precursor |
| 47 | 13 | 3 | 5 | 38.5 | P55290 | Cadherin-13 precursor |
| 48 | 6 | 2 | 4 | 37.0 | P61769 | Beta-2-microglobulin precursor |
| 49 | 5 | 2 | 16 | 36.7 | P11279 | Lysosome-associated membrane glycoprotein 1 precursor |
| 50 | 12 | 2 | 4 | 36.5 | P01625 | Ig kappa chain V-IV region Len |
| 51 | 9 | 2 | 22 | 36.1 | P35754 | Glutaredoxin-1 |
| 52 | 12 | 2 | 11 | 35.6 | P19320 | Vascular cell adhesion protein 1 precursor |
| 53 | 7 | 3 | 2 | 35.4 | P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein |
| 54 | 4 | 3 | 1 | 35.2 | P98164 | Low-density lipoprotein receptor-related protein 2 precursor |
| 55 | 7 | 2 | 1 | 35.1 | P10153 | Nonsecretory ribonuclease precursor |
| 56 | 4 | 3 | 9 | 34.7 | P02751 | Fibronectin precursor |
| 57 | 3 | 2 | 2 | 34.6 | P02774 | Vitamin D-binding protein precursor |
| 58 | 7 | 3 | 6 | 34.1 | P04080 | Cystatin-B |
| 59 | 23 | 3 | 24 | 33.9 | P00441 | Superoxide dismutase [Cu-Zn] |
| 60 | 8 | 2 | 12 | 33.5 | P05090 | Apolipoprotein D precursor |
| 61 | 5 | 2 | 12 | 33.3 | Q15828 | Cystatin-M precursor |
| 62 | 6 | 2 | 14 | 33.1 | P29966 | Myristoylated alanine-rich C-kinase substrate |
| 63 | 4 | 2 | 15 | 32.0 | P31025 | Lipocalin-1 precursor |
| 64 | 4 | 2 | 12 | 31.8 | P78324 | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor |
| 65 | 5 | 2 | 5 | 31.7 | P06870 | Kallikrein-1 precursor |
| 66 | 9 | 2 | 8 | 31.2 | O00468 | Agrin precursor |
| 67 | 4 | 2 | 1 | 29.5 | Q07075 | Glutamyl aminopeptidase |
| 68 | 4 | 2 | 3 | 28.7 | Q96S96 | PEBP family protein precursor |
| 69 | 4 | 2 | 6 | 28.5 | P02765 | Alpha-2-HS-glycoprotein precursor |
| 70 | 5 | 2 | 4 | 28.2 | P11684 | Uteroglobin precursor |
| 71 | 6 | 2 | 12 | 27.6 | Q6GTX8 | Leukocyte-associated immunoglobulin-like receptor 1 precursor |
| 72 | 4 | 2 | 9 | 26.2 | P02766 | Transthyretin precursor |
| 73 | 4 | 2 | 8 | 26.1 | P01040 | Cystatin-A |
| 74 | 4 | 2 | 21 | 26.0 | O75368 | SH3 domain-binding glutamic acid-rich-like protein |
| 75 | 6 | 2 | 20 | 25.2 | P07360 | Complement component C8 gamma chain precursor |
| 76 | 3 | 2 | 7 | 24.2 | P01008 | Antithrombin-III precursor |
| 77 | 4 | 2 | 4 | 24.2 | Q01459 | Di-N-acetylchitobiase precursor |
| 78 | 6 | 2 | 3 | 24.1 | Q13508 | Ecto-ADP-ribosyltransferase 3 precursor |
| 79 | 3 | 2 | 2 | 23.7 | Q14019 | Coactosin-like protein |
| 80 | 6 | 2 | 16 | 23.5 | P16070 | CD44 antigen precursor |
| 81 | 4 | 2 | 1 | 23.2 | P06310 | Ig kappa chain V-II region RPMI 6410 precursor |
| 82 | 3 | 2 | 9 | 23.0 | Q9UM22 | Mammalian ependymin-related protein 1 precursor |
| 83 | 3 | 2 | 8 | 22.8 | Q14315 | Filamin-C |
| 84 | 2 | 2 | 1 | 22.8 | P02735 | Serum amyloid A protein precursor |
| 85 | 11 | 2 | 29 | 22.3 | Q02747 | Guanylin precursor |
| 86 | 4 | 2 | 16 | 21.8 | P02671 | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A] |
| 87 | 6 | 2 | 2 | 21.6 | Q9NP84 | Tumor necrosis factor receptor superfamily member 12A precursor |
| 88 | 4 | 2 | 7 | 21.6 | Q9NQC3 | Reticulon-4 |
| 89 | 6 | 2 | 1 | 21.6 | P07148 | Fatty acid-binding protein, liver |
| 90 | 2 | 2 | 8 | 21.4 | P53634 | Dipeptidyl-peptidase 1 precursor |
| 91 | 4 | 2 | 3 | 21.4 | P02788 | Lactotransferrin precursor |
| 92 | 2 | 2 | 1 | 21.2 | P10619 | Lysosomal protective protein precursor |
| 93 | 2 | 2 | 2 | 21.1 | P08174 | Complement decay-accelerating factor precursor |
| 94 | 4 | 2 | 3 | 20.9 | P01871 | Ig mu chain C region |
| 95 | 18 | 3 | 2 | 20.9 | P01842 | Ig lambda chain C regions |
| 96 | 2 | 2 | 18 | 20.3 | P37173 | TGF-beta receptor type-2 precursor |
| 97 | 2 | 2 | 2 | 20.1 | Q03405 | Urokinase plasminogen activator surface receptor precursor |
| 98 | 4 | 2 | 3 | 19.9 | P09668 | Cathepsin h precursor |
| 99 | 2 | 2 | 3 | 19.8 | P02792 | Ferritin light chain |
| 100 | 2 | 2 | 8 | 19.8 | P36957 | Dihydrolipoyllysine-residue succinyltransferase, mitochondrial precursor |
| 101 | 4 | 2 | 3 | 19.7 | P01703 | Ig lambda chain V-I region NEWM |
| 102 | 5 | 2 | 10 | 19.6 | P10599 | Thioredoxin |
| 103 | 7 | 2 | 12 | 19.0 | P81605 | Dermcidin precursor |
| 104 | 4 | 2 | 10 | 19.0 | P06396 | Gelsolin precursor |
| 105 | 2 | 2 | 1 | 19.0 | P05413 | Fatty acid-binding protein, heart |
| 106 | 6 | 2 | 9 | 18.9 | P01034 | Cystatin-C precursor |
| 107 | 2 | 2 | 7 | 18.7 | P24855 | Deoxyribonuclease-1 precursor |
| 108 | 3 | 3 | 4 | 18.6 | P01700 | Ig lambda chain V-I region HA |
| 109 | 4 | 2 | 11 | 18.3 | A0AVF1 | Tetratricopeptide repeat protein 26 |
| 110 | 5 | 2 | 1 | 18.1 | P05060 | Secretogranin-1 precursor |
| 111 | 2 | 2 | 2 | 18.1 | O75223 | Uncharacterized protein C7orf24 |
| 112 | 2 | 2 | 5 | 18.1 | Q09666 | Neuroblast differentiation-associated protein AHNAK |
| 113 | 6 | 2 | 1 | 17.8 | Q9Y624 | Junctional adhesion molecule A precursor |
| 114 | 3 | 2 | 3 | 17.7 | P08236 | Beta-glucuronidase precursor |
| 115 | 12 | 3 | 1 | 17.5 | O43692 | Peptidase inhibitor 15 precursor |
| 116 | 2 | 2 | 3 | 17.3 | P04207 | Ig kappa chain V-III region CLL precursor |
| 117 | 2 | 2 | 6 | 17.3 | Q86Y38 | Xylosyltransferase 1 |
| 118 | 2 | 2 | 1 | 17.3 | Q13201 | Multimerin-1 precursor |
| 119 | 2 | 2 | 1 | 17.2 | P15289 | Arylsulfatase A precursor |
| 120 | 2 | 2 | 3 | 17.1 | Q16378 | Proline-rich protein 4 precursor |
| 121 | 2 | 2 | 11 | 17.1 | P20142 | Gastricsin precursor |
| 122 | 3 | 2 | 2 | 17.0 | P52758 | Ribonuclease UK114 |
| 123 | 2 | 2 | 7 | 17.0 | P04279 | Semenogelin-1 precursor |
| 124 | 11 | 3 | 3 | 16.5 | Q9GZM5 | Protein YIPF3 |
| 125 | 2 | 2 | 2 | 16.3 | Q9Y4L1 | Hypoxia up-regulated protein 1 precursor |
| 126 | 2 | 2 | 1 | 16.2 | P07711 | Cathepsin L precursor |
| 127 | 2 | 2 | 4 | 15.7 | Q9Y279 | V-set and immunoglobulin domain-containing protein 4 precursor |
| 128 | 2 | 2 | 2 | 15.4 | P01857 | Ig gamma-1 chain C region |
| 129 | 2 | 2 | 6 | 15.4 | Q9BY77 | Polymerase delta-interacting protein 3 |
| 130 | 3 | 3 | 3 | 15.3 | P04433 | Ig kappa chain V-III region VG precursor |
| 131 | 2 | 2 | 7 | 15.2 | Q9UGM3 | Deleted in malignant brain tumors 1 protein precursor |
| 132 | 2 | 2 | 1 | 15.1 | P04430 | Ig kappa chain V-I region BAN |
| 133 | 2 | 2 | 8 | 15.1 | O43653 | Prostate stem cell antigen precursor |
| 134 | 4 | 2 | 15 | 15.1 | Q9UKL3 | CASP8-associated protein 2 |
| 135 | 2 | 2 | 1 | 15.0 | Q15149 | Plectin-1 |
| 136 | 2 | 2 | 1 | 15.0 | Q6UWV6 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 7 precursor |
| 137 | 2 | 2 | 2 | 15.0 | Q12830 | Nucleosome remodeling factor subunit BPTF |
| 138 | 2 | 2 | 1 | 15.0 | P05543 | Thyroxine-binding globulin precursor |
| 139 | 2 | 2 | 2 | 15.0 | P05067 | Amyloid beta A4 protein precursor |
| 140 | 2 | 2 | 1 | 15.0 | P01598 | Ig kappa chain V-I region EU |
| 141 | 2 | 2 | 15 | 15.0 | Q6EMK4 | Vasorin precursor |
| 142 | 2 | 2 | 2 | 15.0 | P02689 | Myelin P2 protein |
TABLE 4.
Summary of Proteins Identified in Human Urine by LC-MS/MS
| Sample Type | Depletion | Protein IDs | Fold |
|---|---|---|---|
| Healthy human urine | No | 29 | — |
| Proteinuria human urine | No | 60 | 2.1 |
| Proteinuria human urine | Yes | 142 | 2.4 |
DISCUSSION
LC-MS/MS is a powerful technique available for proteomic studies. Depending on the type of separation and detection used, reliable detection range may span three to four orders of magnitude of protein concentration. However, this is insufficient for detection of low-abundance proteins that may be present at concentrations up to 10 orders of magnitude lower than the most abundant proteins of the sample. Although the protein content in urine samples of patients with proteinuria is significantly lower compared to plasma, the samples are still very complex and detection of low-abundance proteins remains a difficult task.
The silver-stained SDS-PAGE (Fig. 1) demonstrated efficient depletion of major proteins from human urine. Performance of the affinity depletion MARS column for urine samples was comparable to the performance observed when the column was used for serum samples. In addition, the protocol for removal of abundant proteins from urine required only slight modification, including the removal of low-molecular-weight components using ultrafiltration. Benefits of the commercial depletion kit included efficient removal of targeted proteins and the ability to reuse the column for multiple samples, thus decreasing processing cost per sample.
Importantly, the proteins targeted by the depletion kit (albumin, transferrin, haptoglobin, immunoglobulin G, immunoglobulin A, and alpha-1 antitrypsin) were noticeably absent or greatly reduced in the depleted data set, thus allowing many more moderate- or low-concentration proteins to be found. For example, serum albumin was identified with the highest score in both normal urine (10 peptides) and proteinuria sample (12 peptides), but was identified 20th on the list (only 4 peptides) in the proteinuria sample after depletion.
The human response to disease or infection often increases the number of proteins in the urine. For proteinuria samples, depletion of six high-abundance proteins allowed two-and-a-half times the number of proteins to be identified in urine from these patients. This study demonstrates that depletion is a useful strategy for reducing the overall complexity of the urine sample.
Acknowledgments
This work was supported by the ARUP Institute for Clinical and Experimental Pathology®.
REFERENCES
- 1.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 2.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: An innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 3.Sigdel TK, Lau K, Schilling J, Sarwal M. Optimizing protein recovery for urinary proteomics, a tool to monitor renal transplantation. Clin Transplant. 2008;22(5):617–623. doi: 10.1111/j.1399-0012.2008.00833.x. [DOI] [PubMed] [Google Scholar]
- 4.Darde VM, Barderas MG, Vivanco F. Depletion of high-abundance proteins in plasma by immunoaffinity subtraction for two-dimensional difference gel electrophoresis analysis. Methods Mol Biol. 2007;357:351–364. doi: 10.1385/1-59745-214-9:351. [DOI] [PubMed] [Google Scholar]
- 5.Fu Q, Bovenkamp DE, Van Eyk JE. A rapid, economical, and reproducible method for human serum delipidation and albumin and IgG removal for proteomic analysis. Methods Mol Biol. 2007;357:365–371. doi: 10.1385/1-59745-214-9:365. [DOI] [PubMed] [Google Scholar]
- 6.Thongboonkerd V, Semangoen T, Chutipongtanate S. Enrichment of the basic/cationic urinary proteome using ion exchange chromatography and batch adsorption. J Proteome Res. 2007;6:1209– 1214. doi: 10.1021/pr0605771. [DOI] [PubMed] [Google Scholar]
- 7.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein equalizer technology: The quest for a “democratic proteome. Proteomics. 2006;6:3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 8.Soldi M, Sarto C, Valsecchi C, et al. Proteome profile of human urine with two-dimensional liquid phase fractionation. Proteomics. 2005;5:2641–2647. doi: 10.1002/pmic.200401269. [DOI] [PubMed] [Google Scholar]
- 9.Chromy BA, Gonzales AD, Perkins J, et al. Proteomic analysis of human serum by two-dimensional differential gel electrophoresis after depletion of high-abundant proteins. J Proteome Res. 2004;3:1120–1127. doi: 10.1021/pr049921p. [DOI] [PubMed] [Google Scholar]
- 10.Biesenbach G, et al. [“Diabetic” proliferative retinopathy and nodular glomerulosclerosis without diabetes mellitus] Dtsch Med Wochenschr. 1988;113:1968–1971. doi: 10.1055/s-2008-1067921. [DOI] [PubMed] [Google Scholar]
- 11.Avram MM. Survival in uremia due to systemic diseases. Kidney Int Suppl. 1978:S55–S60. [PubMed] [Google Scholar]
- 12.Burke DG, Emancipator SN, Smith MC, Salata RA. Histoplasmosis and kidney disease in patients with AIDS. Clin Infect Dis. 1997;25:281–284. doi: 10.1086/514556. [DOI] [PubMed] [Google Scholar]
- 13.Prakash J, Singh AK, Saxena RK. Usha. Glomerular diseases in the elderly in India. Int Urol Nephrol. 2003;35:283–288. doi: 10.1023/b:urol.0000020429.14190.5b. [DOI] [PubMed] [Google Scholar]
- 14.van Ypersele de Strihou C, Pirson Y. Indications for dialysis and transplantation in end-stage renal disease. Contrib Nephrol. 1989;71:1104–1110. doi: 10.1159/000417260. [DOI] [PubMed] [Google Scholar]