Abstract
The Staphylococcus aureus plasmid pI258 cadCA operon encodes a P-type ATPase, CadA, that confers resistance to Cd(II)/Pb(II)/Zn(II). Expression is regulated by CadC, a homodimeric repressor that dissociates from the cad operator/promoter upon binding of Cd(II), Pb(II), or Zn(II). CadC is a member of the ArsR/SmtB family of metalloregulatory proteins. The crystal structure of CadC shows two types of metal binding sites, termed Site 1 and Site 2, and the homodimer has two of each. Site 1 is the physiological inducer binding site. The two Site 2 metal binding sites are formed at the dimerization interface. Site 2 is not regulatory in CadC but is regulatory in the homologue SmtB. Here the role of each site was investigated by mutagenesis. Both sites bind either Cd(II) or Zn(II). However, Site 1 has higher affinity for Cd(II) over Zn(II), and Site 2 prefers Zn(II) over Cd(II). Site 2 is not required for either derepression or dimerization. The crystal structure of the wild type with bound Zn(II) and of a mutant lacking Site 2 was compared with the SmtB structure with and without bound Zn(II). We propose that an arginine residue allows for Zn(II) regulation in SmtB and, conversely, a glycine results in a lack of regulation by Zn(II) in CadC. We propose that a glycine residue was ancestral whether the repressor binds Zn(II) at a Site 2 like CadC or has no Site 2 like the paralogous ArsR and implies that acquisition of regulatory ability in SmtB was a more recent evolutionary event.
Numerous cellular processes require metal ions as cofactors for enzymatic reactions or structural components of proteins. Some required metals are toxic at high concentrations, and other metal ions play no physiological roles and are toxic at low concentrations. Therefore, cells have evolved detoxification systems for a variety of metals including arsenic, antimony, zinc, lead, and cadmium (1, 2). These systems are extremely important to the survival of bacteria and are generally composed of a single transcriptionally regulated operon that encodes at least two gene products; that is, a transcriptional repressor (metal sensor protein) and a resistance protein such as an efflux pump, intracellular chelator, or modifying enzyme. The cadCA operon of Staphylococcus aureus plasmid pI258 confers resistance to Cd(II), Pb(II), and Zn(II) and encodes two proteins: CadC, a metal-responsive transcriptional repressor that responds to Cd(II), Pb(II), and Zn(I) (3–5) and CadA, a P-type ATPase that extrudes the soft metals out of the cell (6). The CadC homodimer binds to the cad operator/promoter (o/p)3 and represses transcription of the operon. When CadC binds Cd(II), Zn(II), or Pb(II), it dissociates from the DNA, allowing expression of the CadA ATPase.
CadC has two different types of metal binding sites (four binding sites per dimer) (7, 8). The regulatory site, termed Site 1, is composed of four cysteine resides, Cys7, Cys11, Cys58, and Cys60 (of which Cys11 is not absolutely required) (5). The crystal structure of apo-CadC revealed a second type of binding site, termed Site 2, at the C-terminal interface between the dimers is composed of four residues, Asp101 and His103 from one monomer and His114 and Glu117 from the other monomer (9).
The objective of this study was to examine the roles of Site 1 and Site 2 in CadC function and to shed light on the evolution of these sites. Although Site 2 is not regulatory in CadC, it is congruent with the regulatory Zn(II) site of the homologous SmtB repressor (10). In the crystal structure of the aporepressor, one Site 2 is filled with Zn(II), and the other Site 2 is empty (9). In this study the Site 1 and Site 2 metal binding sites were eliminated by mutagenesis. The stoichiometry, relative affinities, and coordination geometry of both sites of were determined. Both types of sites bound both Cd(II) and Zn(II), but Site 1 had a preference for Cd(II) over Zn(II), and the reverse was found for Site 2. Although mutagenesis of the metal binding residues in Site 2 eliminated binding of Zn(II) at the dimer interface, there was only a minimal effect on dimerization. The structure of the mutant lacking Site 2 was compared with the wild type with bound Zn(II) and with the structures of SmtB with and without Zn(II) (11). The results suggest that a single residue at position 87 may be responsible for the difference between the non-regulatory Zn(II) site in CadC and the Zn(II)-responsive site in SmtB.
MATERIALS AND METHODS
Construction of Mutants and Purification of CadC—CadC with a C11G substitution has wild type metalloregulatory properties and is considered as the wild type for the purposes of this study (5). This variant has improved solubility properties and is less prone to form intermolecular disulfide bonds than CadC with a cysteine residue at position 11. Purified CadC was prepared from Escherichia coli strain BL21(DE3) pYSCM2, as described previously (9). CadC was reduced with 10 mol eq of dithiothreitol at 4 °C for 3–4 h. Dithiothreitol removal and concentration of protein were performed by dialysis using 10-kDa cut dialysis tubing membrane in an anaerobic chamber at 4 °C overnight. The stoichiometry of reduced thiols in purified wild type CadC was determined by titration with 5,5′-dithiobis(2-nitrobenzoic acid) (12) to be 4.0 ± 0.2, consistent with Cys7, Cys52, Cys58, and Cys60 all being fully reduced.
To mutate Site 1, Cys7, Cys58, and Cys60 were altered to serine residues. To mutate Site 2, Asp101 was changed to glycine in a previously constructed H103A mutant (7, 8). Mutants were prepared from the C11G variant using a QuikChange site-directed mutagenesis kit (Stratagene) and were purified by the same procedure as described above.
Fluorescence Anisotropy; DNA Binding by Fluorescence Anisotropy—DNA binding studies by fluorescence anisotropy were made using a Photon Technology International spectrofluorimeter fitted with polarizers in the L format. Complementary 34-mer oligonucleotides, one of which was labeled at the 5′ end with carboxyfluorescein, were synthesized (Integrated DNA Technologies, Inc.) to include the cad operator/promoter regions (5, 13): 5′-carboxyfluorescein-ATAATACACTCAAATAAATATTTGAATGAAGATG-3′ and 3′-TATTATGTGAGTTTATTTATAAACTTACTTACTAC-5′. The two strands were heated to 94 °C for 5 min, annealed by cooling to room temperature, and stored in aliquots at -20 °C. Anisotropy measurements were performed in a buffer consisting of 5 mm MOPS-NaOH, pH 7.0, containing 0.4 m NaCl, 1 mm dithiothreitol, and 5 mm EDTA. Dithiothreitol and EDTA were excluded for metal titrations. The buffer was degassed by bubbling with argon before the assay. Apo or metallated CadCs were titrated with 1.7 ml of 50 nm fluorescein-labeled cad o/p DNA. CadC was metallated by prior incubation with the addition of a mol eq of CdCl2. Changes in anisotropy were calculated after each addition using the supplied Felix32 software. The stoichiometry of binding of the CadC dimer to the cad o/p was calculated using DynaFit (14).
Inductively Coupled Plasma Mass Spectrometry (ICP-MS)—All metal binding experiments were carried out anaerobically under argon at 4 °C. For metal titrations, CadC in a buffer consisting of 25 mm MOPS, pH 7.0, 0.3 m NaCl was mixed with various concentrations of Zn(II) or Cd(II) at the indicated molar ratios in the same buffer. The final volume of each tube was adjusted to 0.1 ml with buffer and incubated 15 to 20 min on ice. Unbound metal was removed using Bio-Gel P-6 Micro Bio-Spin columns (Bio-Rad). For experiments in which bound Cd(II) was replaced by Zn(II), 2 mol eq of ZnCl2 were added to CadC before the Cd(II) titration. The metal content of eluted samples was determined by ICP-MS using a PerkinElmer Life Sciences ELAN 9000. All samples were dissolved in 2% analytical grade HNO3 acid for analysis. Standard solutions were purchased from PerkinElmer Life Sciences for calibration. The protein concentration was determined using a calculated molar extinction coefficient at 280 nm of 6585 m-1 cm-1 (15) or a Bio-Rad protein assay using bovine serum albumin as a standard.
X-ray Absorption Spectroscopy—Samples for XAS analysis were prepared under argon in a buffer containing 25 mm MOPS-KOH, pH 7, 0.3 m NaCl, and 30% (v/v) glycerol Zn(II) or Cd(II) were added at either 0.9 or 1.8 mol eq to fill one or both types of sites. In experiments with both cadmium and zinc, the two metals were each added at 0.9 mol eq. Samples were added to Lucite cuvettes and frozen in liquid nitrogen.
Zinc and cadmium XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamlines 9-3 and 10-2 and at the National Synchrotron Light Source (NSLS) on beamline X9b. SSRL beamlines 9-3 and 10-2 were equipped with Si111 double crystal monochromators; beamline 9-3 utilized a mirror for harmonic rejection, whereas 10-2 was detuned 50% at k = 13 Å-1 at each element. NSLS beamline X9b was equipped with a Si111 monochromator equipped with a harmonic rejection mirror. SSRL samples were maintained at 10 K using Oxford Instruments continuous-flow liquid helium cryostats, and NSLS samples were run at 24 K using a He Displex Cryostat. Protein fluorescence excitation spectra were collected using 30-element Ge solid-state array detector at SSRL beamline 9-3 and using 13-element GE Healthcare solid-state detectors at both SSRL beamline 10-2 and at NSLS beamline X9b. X-ray energies were calibrated by collecting zinc or cadmium foil absorption spectra simultaneously with protein data, assigning the first inflection point as 9659 eV for zinc and 26725 eV for cadmium. Each fluorescence channel of each scan was examined for spectral anomalies before averaging. SSRL 9-3 data represent the average of 5 to 6 scans, whereas SSRL 10-2 and NSLS data represent the average of 9–10 scans. Full XAS data sets were collected on multiple independent wild type and mutant CadC samples to ensure sample reproducibility.
XAS data were processed using the Macintosh OSX version of the EXAFSPak program suite integrated with the Feff v7.0 software for theoretical model generation. Data reduction was performed following preexisting protocols (16). Data were converted to k-space using an E0 = 9680 eV for zinc and 26730 eV for cadmium. The k3-weighted EXAFS was truncated between 1.0 and 13 Å-1 for both metals for filtering purposes and Fourier-transformed. EXAFS data-fitting analysis was performed on raw/unfiltered data. Single scattering Feff Version 7.0 models were calculated for zinc- and cadmium-carbon, oxygen, sulfur, and zinc/cadmium coordination scattering to simulate possible metal-ligand environments. During data simulation, only the bond length and Debye-Waller factors were allowed to freely vary, whereas coordination numbers were fixed at half-integer values. For zinc fits, values for the scale factor of 1.0 and an E0 of -15.25, determined from fitting crystallographically characterized models, were fixed and utilized in the protein spectral simulations (16). For cadmium fits, values for the scale factor of 1.0 and an E0 of -10.00, determined from fitting crystallographically characterized models, were fixed and utilized in the protein spectral simulations. Criteria for judging the best fit simulation and for the addition of ligand environments to the fit included the lowest mean square deviation between data and fit (F′), a value that was corrected for number of degrees of freedom in the fit, and all Debye-Waller factors maintained below a maximum value of 0.006 Å2. Simulation parameters of the progressively optimized zinc and cadmium EXAFS data for each sample are in supplemental Tables 1 and 2.
Isothermal Titration Calorimetry (ITC) Assays—Protein and aqueous ZnCl2 solutions were prepared anaerobically in 25 mm MOPS, pH 7.0, containing 0.3 mm NaCl. ITC experiments were conducted anaerobically at 25 °C using a VP-ITC titration microcalorimeter (MicroCal, Inc., Northampton, MA) by titrating a 4 mm metal solution into 0.2 mm CadC in 1.4 ml of buffer. After an initial 2 μl of ZnCl2 solution injection, 40 additional injections of 3 μl each were titrated into the protein sample at 5-min intervals. The syringe stirring speed was held constant at 500 rpm. All experiments were conducted in triplicate on independent protein and ZnCl2 samples to ensure data reproducibility. Data analysis was performed utilizing Origin 7.0 Scientific Graphing and Analysis Software (MicroCal) by applying a nonlinear least-squares curve-fitting algorithm.
Optical Spectroscopy Measurements of Cd(II) Binding by CadC—UV-visible absorption measurements were carried out under argon at room temperature in a Varian Carry 1E spectrophotometer. Aliquots of CdCl2 were added from a gastight Hamilton syringe to reduced CadC in a sealed cuvette. EDTA was used to buffer very low concentrations of free Cd(II). A conditional stability constant for the Cd-EDTA complex was 3.2 × 1012 m-1 under these experimental conditions (17). After each addition the solution was mixed and allowed to equilibrate for 2 min, after which spectra were collected from 200 to 600 nm. Spectra were corrected for dilution and by subtracting the apoCadC spectrum from each spectrum of metallated protein.
Ultracentrifugation Analysis of CadC—Wild type CadC and the Site 2 mutant were analyzed by sedimentation velocity ultracentrifugation at the National Analytical Ultracentrifugation Facility at the University of Connecticut.
Crystallization of the Site 2 Mutant—Crystals of the Site 2 mutant were grown at 23 ± 2 °C in hanging drops. Purified protein (2 μl of 13.5 mg/ml) was added to 2 μl of a well solution containing 1.6 m (NH4)2SO4 and 0.1 m sodium citrate, pH 5.0. The droplets were equilibrated with 0.6 ml of well solution. The crystals grew within 4–5 days, attaining dimensions of 0.3 × 0.15 × 0.15 mm3. Using a well solution containing 30% glucose, data were collected at the Advance Photon Source, LS-CAT Beamline 21-ID-D, Chicago, IL. The crystal belonged to the space group P43, with cell parameters a = b = 89.47 Å, c = 148.54 Å, α = β = γ = 90°. The structure was solved by molecular replacement using Phaser (24) using a dimer from the CadC crystal structure (PDB code 1U2W) as a model (9). The solution was further refined by the maximum likelihood method using REFMAC (18) of the CCP4 suite (19). A total of 142 water molecules were identified in the 2Fo - Fc map. A few peaks that appeared at >3.5 σ in the difference Fourier map were identified Na+ ions. Attempts to model zinc atoms with partial occupancy into those made the R- and B-factors worse. There were a few unexplained densities in the final difference Fourier map. The final R-factor after refinement to convergence was 28.8%, with a free R value of 28.6%. X-ray data and refinement statistics are in supplemental Table 1.
RESULTS
Construction of Site 1 and Site 2 Mutants—Mutant CadCs were created that eliminated either Site 1 or Site 2. To the Site 1 mutant, Cys7, Cys58, and Cys60 were sequentially altered to serine residues. In this mutant Site 2 is unaltered. To construct the Site 2 mutant, Asp101 was changed to glycine residue in a previously constructed H103A mutant (7, 8). This double mutant was used because, as described below, it crystallized well. The wild type and two mutants were purified, and their DNA and metal binding properties were compared.
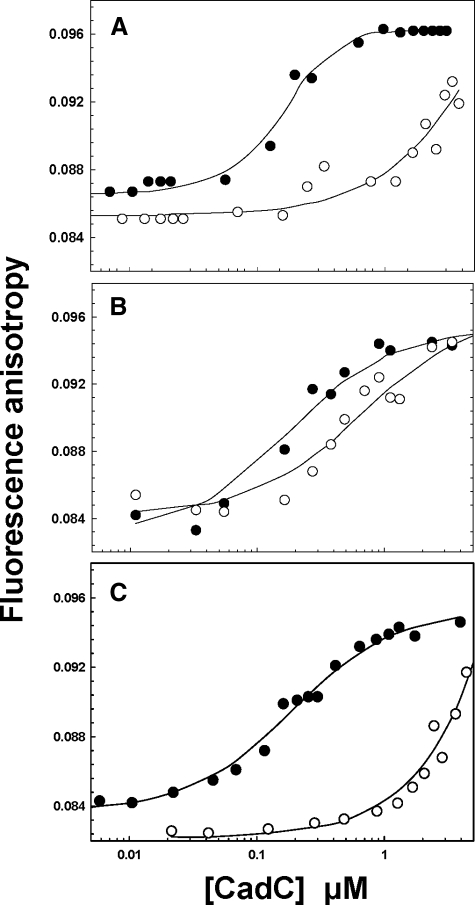
DNA Binding and Cd(II)-dependent Dissociation—To examine the effect of the two types of mutations on binding to the cad o/p, fluorescence anisotropy experiments were performed using a 5′-fluorescein-labeled 34-bp double-stranded oligonucleotide containing the cad o/p. Binding was estimated from the change in fluorescence anisotropy (Fig. 1). The magnitude of the binding affinities reveals that the three apoproteins bind to DNA with similar affinities. Wild type CadC has only an ∼2-fold higher affinity (Ka = 1.3 ± 0.02 × 107 m-1) (Fig. 1A) than the Site 1 (6.4 ± 0.01 × 106 m-1) (Fig. 1B) and Site 2 mutants (Ka = 6.2 ± 0.01 × 106 m-1) (Fig. 1C). Binding of Cd(II) to wild type CadC reduces the affinity for DNA by nearly 2 orders of magnitude relative to the apoprotein (Fig. 1A). Similarly, binding of Cd(II) to the Site 2 mutant decreased the affinity for DNA approximately by more than 2 orders of magnitude compared with its apo form (Fig. 1C), demonstrating that the Site 2 mutant retains near wild type response to Cd(II). In contrast, Cd(II) had little effect on the affinity of the Site 1 mutant for DNA (Fig. 1B), indicating loss of metalloregulation of DNA binding by elimination of Site 1.
FIGURE 1.
Wild type, Site 1, and Site 2 mutant CadCs binding to and Cd(II)-induced dissociation from cad o/p DNA. Protein-DNA interactions were monitored by fluorescence anisotropy using a 34-bp double-stranded oligonucleotide labeled on one strand with 5′-fluorescein. Binding isotherms were generated by titration of unmetallated (•) or metallated (○, A), wild type (B), Site 1 mutant, and Site 2 mutant (C).
Analysis of Binding of Zn(II) and Cd(II) to Wild Type and Mutant CadCs by ICP-MS—The binding properties of wild type CadC has been characterized by optical spectroscopy for Cd(II) and Pb(II), which absorb light at ∼240 nm when they form a charge-transfer intermediate with the cysteine thiolates of Site 1 (8). Zn(II), however, does not produce a spectroscopic signal upon binding in CadC Site 1 nor do the metal-O and metal-N bonds in Site 2 produce a detectable absorbance.
On the other hand, high affinity metal binding can be determined directly by ICP-MS, with which multiple metals can be analyzed simultaneously. Wild type CadC and the Site 1 and Site 2 mutants were titrated with either Cd(II) or Zn(II) (supplemental Fig. 1). Wild type protein bound Zn(II) with a stoichiometry of 4 per CadC dimer but only two Cd(II) per dimer (supplemental Fig. 1A), indicating four binding sites for Zn(II) but only two for Cd(II). The Site 2 mutant, which retains Site 1, bound either Zn(II) or Cd(II) in a ratio of two metals per dimer (supplemental Fig. 1B), suggesting that Site 1 binds either metal. Surprisingly, the Site 1 mutant, which retains Site 2, also bound two atoms of either Zn(II) or Cd(II) (supplemental Fig. 1B), indicating that Site 2 also binds either metal.
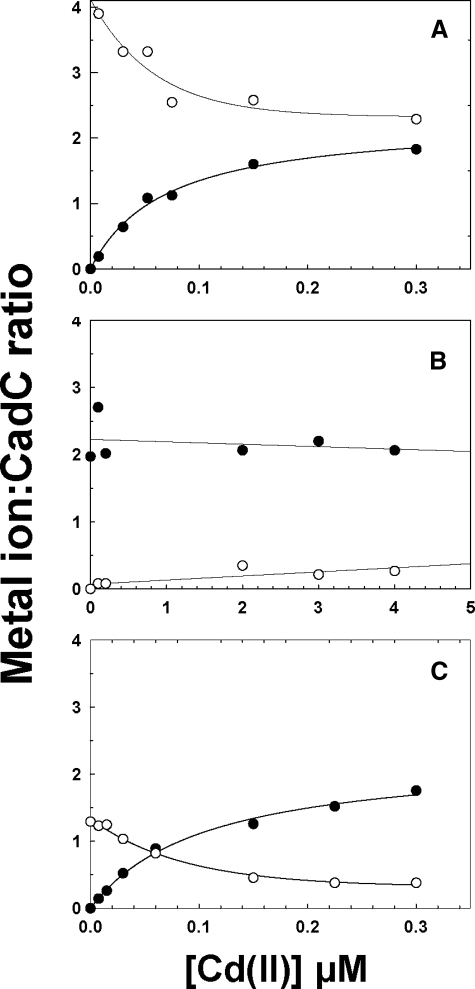
Which sites bind which metals? To investigate this question, wild type CadC was saturated with four Zn(II) atoms, and then the Zn(II)-bound protein was titrated with Cd(III) (Fig. 2A). Using ICP-MS, the same sample could be assayed for both metals simultaneously. At saturating Cd(II), two of the four Zn(II) atoms were displaced, resulting in a CadC dimer with two Cd(II) and two Zn(II) atoms. These results indicate that one type of site is selective for Zn(II) over Cd(II), and the other has higher affinity site for Cd(II) than Zn(II). In the Site 2 mutant, Zn(II) was displaced by Cd(II) (Fig. 2C). In contrast, in the Site 1 mutant Zn(II) was not displaced by Cd(II) (Fig. 2B). These results suggest that Site1 is selective for Cd(II) over Zn(II), whereas Site 2 is selective for Zn(II) over Cd(II).
FIGURE 2.
Cd(II) displaces Zn(II) in Site 1 but not Site 2. Wild type (A), Site 1 mutant (B), and Site 2 mutant (C) CadCs were metallated with a 2–3-fold excess of Zn(II) per dimer and then titrated with the indicated concentrations of Cd(II). The amount of Zn(II) (○) and Cd(II) (•) bound was determined in the same sample by ICP-MS. In the wild type all four sites were initially filled with Zn(II), and Cd(II) displaced two per dimer. The mutants both bound only two Zn(II) per dimer. In the Site 1 mutant, the Zn(II) could not be displaced by Cd(II), but it was displaceable in the Site 2 mutant.
Cd(II) binding isotherms for wild type and mutant CadCs were calculated from the amount of bound Cd(II) as a function of the [Cd(II)]/[CadC] molar ratio using Dynafit (14) (data not shown). The fit of the data shown is consistent with binding of two cadmium atoms per dimer for all three proteins, indicating that each has single type of tight Cd(II) binding site, in agreement with EXAFS data described below. The sensitivity of the method allows estimation of only a lower limit of Kd (1 > 107 m-1) under these experimental conditions. The affinity of the wild type and Site 2 mutant was too high for determination, but affinity of the Site 1 mutant (Kd = 4.3 ± 0.01 × 106 m-1) could be calculated. The results show that only two cadmium atoms are bound with high affinity in the wild type and Site 2 mutant, i.e. to Site 1. Thus, although both Site 1 and Site 2 appear individually capable of binding Cd(II), Site 2 binds only low affinity.
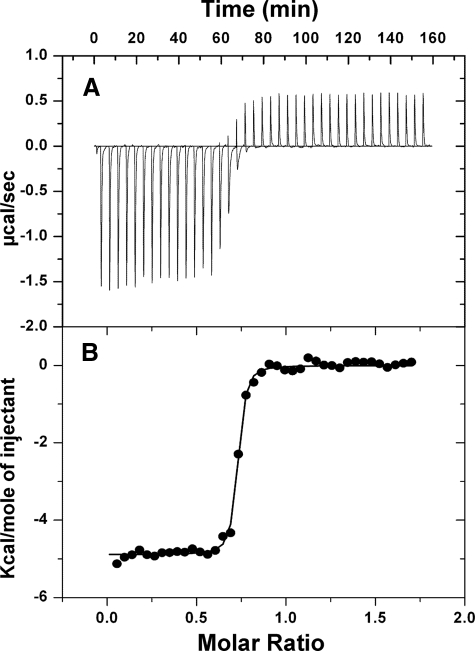
Binding of Cd(II) to Site 2 Assayed by Isothermal Titration Calorimetry—To confirm that Cd(II) binds to Site 2 at low affinity, binding was assayed by ITC. Again, the binding affinity for wild type and the Site 2 mutant, i.e. at Site 1, is too high to be determined accurately by this method, but the affinity of the Site 1 mutant, i.e. binding of Cd(II) at Site 2, was within the range of ITC analysis (Fig. 3). The data displayed a mixed heat reaction that began with exothermic and was followed by late exothermic heat changes. Cd(II) binding follows an exponential binding curve in the initial stages followed by constant heat absorption due to Cd(II) hydration at the post-saturation sate. The best fit binding isotherm for Cd(II) binding was found to have a stoichiometry of 1.4 ± 0.02 per dimer, consistent with the value of 2 cadmiums per dimer found by the ICP-MS direct binding measurements. The Ka for Cd(II) was determined to be 8.1 ± 1.4 × 106 m-1, which is in the same range as binding affinity observed (Kd = 4.3 ± 0.01 × 106 m-1) from the direct binding measurements by ICP-MS.
FIGURE 3.
Cd(II) binds to Site 2 with low affinity. A, isothermal titration calorimetry of Cd(II) binding to the Site 2 mutant. B, the area under each injection signal was integrated, and the enthalpy per mol of Cd(II) injected was calculated as Cd(II)/protein. The solid line represents a nonlinear least-squares fit of the reaction heat per injection calculated with Origin 7.0 Scientific Graphing and Analysis Software from MicroCal.
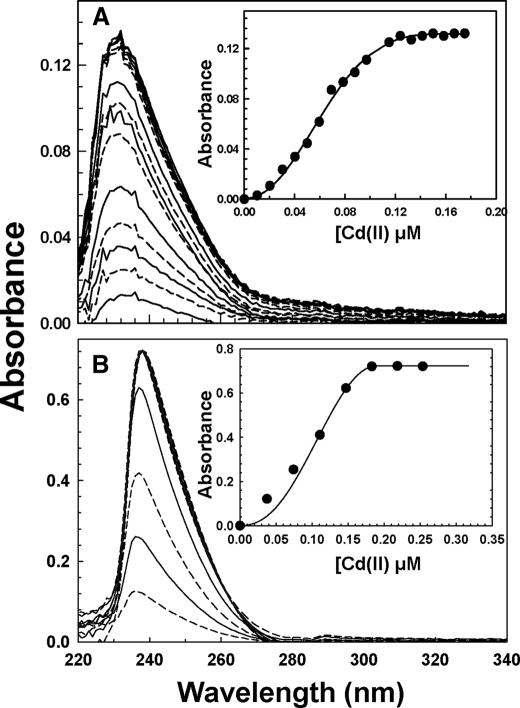
Binding of Cd(II) to Site 1 by Optical Spectroscopy—The intense cadmium-thiolate charge-transfer band at ∼240 nm is useful for determining the relative affinities of Site 1 and Site 2. Wild type and mutant CadCs were titrated with Cd(II) in the presence of EDTA (Fig. 4). Both wild type (Fig. 4A) and the Site 2 mutant (Fig. 4B) produce a peak centered at 238 nm. The association constants of Cd(II) binding were calculated using DynaFit (14) with assumed stoichiometries of 1:1, 2:1, or 4:1 Cd(II):CadC dimer. Assumptions of 1:1 and 4:1 stoichiometries gave unreasonably high standard errors, whereas the 2:1 stoichiometry was within acceptable limits. Association constants of 4.1 × 1012 and 2.6 × 1012 m-1 were calculated for the wild type and Site 2 mutants, respectively, demonstrating that Site 1 is a high affinity cadmium binding site.
FIGURE 4.
Cd(II) binds to Site 1 with high affinity. Each curve represents the ultraviolet spectrum of an anaerobic sample of wild type (A) or Site 2 mutant (B) with Cd(II). The curves alternate between solid and dashed lines, indicating increasing concentrations of Cd(II), as indicated in the insets, which show the Cd(II) binding isotherms.
Analysis of Cadmium and Zinc Binding by X-ray Absorption Spectroscopy—X-ray absorption spectroscopy was used to probe the local structural environment of zinc and cadmium metal bound to the individual CadC metal binding sites. Full zinc EXAFS were collected for a number of samples including 1) wild type CadC with two or four zincs bound per protein dimer, 2) wild type protein with both two zinc and two cadmium bound per dimer, 3) the Site 1 mutant with two zinc bound per dimer, and 4) the Site 2 mutant with two zinc bound per dimer (supplemental Fig. 2). Full cadmium EXAFS were collected on the complementary set of samples. Raw and simulated zinc and cadmium EXAFS for each sample are given in spectra A–E in supplemental Figs. 2 and 3, respectively. Corresponding Fourier transforms of the raw and simulated EXAFS are given in spectra F–J in supplemental Figs. 2 and 3.
In the case of the wild type samples with two or four zincs bound per dimer in the absence of an additional metal, spectral simulations indicate nearly identical average zinc coordination environments between the two samples constructed on average of 2.0 O/N and 1.5 S nearest neighbor zinc ligands at respective average distances of 1.97 and 2.20 Å (supplemental Table 2, fits 1.4 and 2.4). In both cases, additional long range Zn···C scattering is observed at ca. 3 and 4 Å. The strong similarity between the best-fit averaged zinc-nearest neighbor bond parameters for the wild type, regardless of whether two or four zinc atoms are bound per dimer suggests that in the absence of additional metals, zinc populates both Sites 1 and 2 equally. Upon the addition of stoichiometric amounts of cadmium and zinc to the wild type, the average zinc environment becomes constructed of only oxygen/nitrogen-based ligands (there is no Zn-S scattering present in this sample). The initial 1.97 Å Zn-O/N ligand shell seen in the zinc-only wt data is resolved at 1.95 Å in the mixed metal wt-sample; in addition, a second resolvable Zn-O/N nearest neighbor environment at 2.09 Å was also observed (supplemental Table 2, fit 3.3). The appearance of the additional resolvable Zn-O/N shell may reflect subtle perturbations in Site 2 structure upon cadmium binding at Site 1 that makes this interaction observable in the mixed metal species. Alternatively, a portion of the metal may bind at an adventitious metal binding site, or free metal in water should also account for this new site. Regardless of its origin, the average nearest neighbor coordination number increases in the mixed metal sample, suggesting a higher average coordination state for bound zinc. Long range Zn···C scattering for this sample is again seen at ∼4 Å. These results indicate that when zinc is present by itself, it fills all four sites, but when both metals are present, zinc fills only the two Site 2, and cadmium prefers the regulatory Site 1.
To examine the selectivity of the two sites in more details, zinc XAS experiments were also performed on the Site 1 and Site 2 mutants. As seen in samples of wild type protein with both metals present, zinc EXAFS for that Site 1 mutant are best simulated with only oxygen/nitrogen based; there is again no evidence for sulfur ligation in this mutant sample. However, unlike the wild type sample with both metals, the nearest neighbor environment in the Site 1 mutant could be simulated only with a single Zn-O/N ligand environment at an average bond distance of 1.96 Å. Long-range scattering was consistent again with carbon scattering at ∼4 Å. Fitting analysis for the Site 2 mutant indicated that nearest neighbor scattering was dominated in this sample by Zn-S ligation at 2.31 Å (supplemental Table 2, fit 5.4). Additional Zn-O/N scattering at 1.94 and 2.08 Å is also observed. Because Site 2 is absent, this may represent adventitious binding again at a nonspecific site or free metal in solution. The presence of Zn···C scattering at ∼4Å was also observed in this sample, suggesting a portion of the metal is bound to the protein.
Complementary EXAFS experiments were performed on wild type and mutant CadC with cadmium. Simulations of the cadmium EXAFS for wild type CadC with two cadmium atoms bound/dimer in the absence of additional metals are best fit with a ligand environment of 2.5 Cd-O/N and 2.0 Cd-S atoms at 2.30 and 2.50 Å, respectively (supplemental Table 3, Fit 6.3). Long range (R > 3 Å) signals are best fit with only carbon scattering at 3.25 Å. Upon the addition of a second equivalent of cadmium to this sample, there is subtle rearrangement of the averaged cadmium environment, with the best-fit showing 1.5 Cd-O/N atoms at 2.31 Å and 2.5 Cd-S atoms at 2.52 Å (supplemental Table 3, fit 7.3). Long-range scattering is best fit with carbon-only scattering at 3.70 Å. The addition of 2 zinc atom/protein dimer to wild type CadC with 2 cadmiums bound is best fit with 2.0 Cd-O/N ligands at 2.31 Å and 3.0 Cd-S ligands at 2.51 Å (supplemental Table 3, fit 8.3). Long range Cd···C scattering was observed in this sample at 3.83 Å. In the case of the Site 1 mutant, the cadmium-nearest neighbor environments are best fit with only oxygen/nitrogen-based ligands; there is no evidence for sulfur ligation in this sample. Two unique resolvable Cd-O/N interactions are observed in the regulatory site mutant at 2.25 Å (coordination number of 3) and at 2.39 Å (coordination number of 2). Long-range carbon scattering was observed at 3.22 Å in this sample. In the case of the Site 2 mutant, the cadmium nearest neighbor environment is constructed of 2 Cd-S interactions at 2.52 Å and 2 Cd-O/N interactions at 2.29 Å. Long-range scattering was observed in this case at 3.79 Å. These results are most consistent with Cd(II) filling Site 1 in both the wild type and Site 2 mutant with some free cadmium with bound solvent.
Site 2 Is Not Required for Dimerization of CadC—Analysis of the data from sedimentation velocity ultracentifugation of the two CadCs showed that both samples behaved as single species with sedimentation coefficients, s20,w, of 2.13 and 2.16 S, respectively (data not shown). The buoyant molecular masses are in good agreement with those expected for dimers of each protein, with no evidence of dissociation of the dimers or further association. Under the experimental conditions used, a reversible monomer-dimer self-association could have been detected if the Kd were 300 nm or greater. Thus, over the concentration range studied, both wild type and mutant CadCs behaved as tight, non-dissociating dimers, demonstrating that zinc binding at Site 2 has no effect on for dimerization.
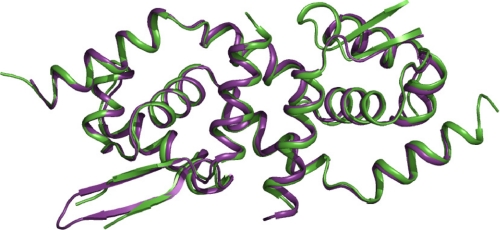
Crystal Structure of the Site 2 Mutant—The crystal structure of the Site 2 mutant was solved and refined to 2.31Å (PDB code 3F72). The structure contains three molecules in the asymmetric unit, termed dimers AB, CD, and EF. The root mean square deviations in the positions of the common atoms among the three dimers as well as the wild type structure were less than one. From superposition of dimer CD with the wild type structure in which zinc was present in Site 2, it is apparent that the presence or absence of zinc makes no substantial difference to the overall structure (Fig. 5). A few residues from the N and C termini were missing in the electron density map of the Site 2 mutant as well as in the loop region spanned by residues 89–96. The disorder in those regions, which represent the main differences between the dimers, could not be resolved. A few residues were missing side chain densities and were modeled as alanines. The average B-factor of the structure is 56.5. Residues in the regions of disorder had higher B-factors.
FIGURE 5.
Superposition of the zinc-bound and zinc-free forms of CadC. Crystal structure of CadC Site 2 mutant (green) superposed on that of wild type CadC with Zn(II) bound in both Site 2s (purple).
From the electron density map, it is clear that there is no bound zinc at the position of the altered Site 2. The only non-protein densities were observed at >3.5 σ contour of the Fo - Fc map at a region closer to His114 of dimer AB and 2.26 Å away from the corresponding positions of the zinc atoms in the wild type structure. Further refinement identified the densities as Na+ ions. Of the five Na+ identified from the electron density map, four were bound to dimer AB, whereas the fifth was bound to dimer EF. Dimer CD was devoid of cations and was used as for further analysis, but the results were essentially the same with the other two dimers. Thus, the D101G and H103A substitutions eliminated binding of Zn(II) at Site 2 with no major changes to the overall structure.
DISCUSSION
In this report we examined the role of the two types of CadC metal binding sites, Site 1, the regulatory site, and Site 2, present at the interface between monomers. We focused on three questions; 1) what is metal selectivity of the two sites. 2) what role, if any, does nonregulatory Site 2 play in CadC structure or function. and 3) what differentiates the nonregulatory Site 2 in CadC from the homologous Zn(II) regulatory Site 2 in SmtB, and what does that imply for the evolution of this site?
First, the two types of sites were found to have different selectivity, with Site 1 preferring Cd(II) and Site 2 preferring Zn(II). Both sites could be filled with Zn(II), but the addition of Cd(II) displaced only the two zinc atoms in Site 1, not the two in Site 2. This is not surprising as Site 1 has three or four cysteine thiolates that prefer the softer metal Cd(II). Site 2, with two imidazole nitrogens and two carboxylate oxygens, would be expected to prefer the harder metal Zn(II). Although Cd(II) binding to Site 2 was not observed in wild type CadC when assayed by direct binding assays using ICP-MS, this probably represents the approximately 6 orders of magnitude difference in affinity between Site 1 and Site 2 for cadmium. Cd(II) binding to Site 2 is only observed when Site 1 is absent, i.e. in the Site 1 mutant (supplemental Fig. 1B).
The second question is, Does Zn(II) binding to Site 2 have a structural or functional role in CadC? It is not regulatory. The Site 1 mutant loses all regulatory activity in vivo (5), and Cd(II) does not induce dissociation from the cad o/p in vitro (Fig. 1B). In contrast, there seems to be little effect of removing the ability of Site 2 to bind Zn(II) or to dissociate from the DNA upon the addition of inducer (Fig. 1C). Neither does the presence of Zn(II) in Site 2 have an obvious structural role, as sedimentation equilibrium analysis indicates that both wild type and the Site 2 mutant of CadC are equally tight dimers. Comparison of the structure of wild type with zinc bound at Site 2 and the Site 2 mutant with no zinc shows no significant differences (Fig. 5), so the presence of zinc appears to make little difference in CadC. Therefore, at this time we cannot assign a role for Site 2.
That brings up the third topic; What is the difference between Zn(II) binding to Site 2 in CadC and the corresponding regulatory Zn(II) binding site in the Zn(II)-responsive repressor SmtB? The ArsR/SmtB family of metalloregulatory proteins have a common DNA binding helix-turn-helix fold with a winged helix structure. In the absence of their inducing metal ions, these proteins bind to the promoter region of their respecting operons, thus repressing transcription. When the inducing metal is bound, the repressor dissociates, presumably as a result of a conformational change in the DNA binding region, thus allowing for transcription. Although the overall fold in these family members is conserved, the binding sites for inducer metals in different homologues are located at a number of locations on the surface of the protein, apparently by convergent evolution (20). Presumably, metal binding at each of these disparate sites transmits a conformational change to the DNA binding site; it is not necessary that they each cause the same change as long as the metallated repressor no longer binds to the DNA with the same affinity.
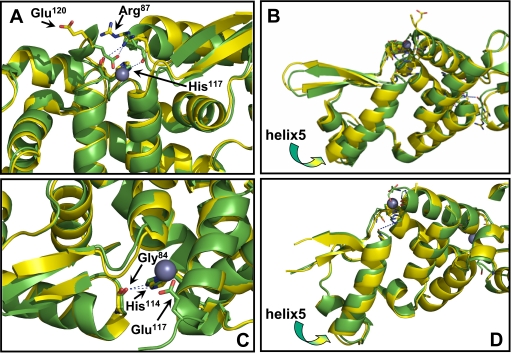
CadC and SmtB have a sequence similarity of 48.4% and a 79% structural identity (21). Crystal structures have been solved for both CadC (9) and SmtB (22). The root mean square deviation of the position of the common atoms between the two structures is 2.26 Å. In both, a helix-turn-helix interacts with the DNA. Comparison of apo and zinc-bound forms of SmtB indicate that a series of interactions propagating from Site 2 affects the DNA binding region (11). These include dislocation of His117 and Glu120 toward the binding site, changes in the backbone hydrogen bonding distances between Leu83 and Leu88, and the interactions made by Arg87. The backbone of Arg87 forms hydrogen bonds with side chain His117 when zinc binds to SmtB. In addition, the side chain of Arg87 forms hydrogen bonds with the side chain of Glu120 and the backbone carboxyl group of His117 (Fig. 6A). Changes in the conformation of Arg87 influence DNA binding because it is positioned at the C terminus of the first helix of the DNA binding site (Fig. 6B). In CadC, the residues corresponding to His117 and Glu120 are His114 and Glu117. These show similar dislocation toward the zinc binding site in the zinc-bound form of CadC. The CadC residue corresponding to SmtB Arg87 is Gly84 (Fig. 6C). Interactions of Gly84 with His114 do not create any significant changes in the orientation of CadC helix 5 (Fig. 6D), and thus, binding of Zn(II) to Site 2 site is non-regulatory in CadC. We, thus, propose that Arg87 in SmtB is the key residue that confers regulatory activity in this repressor, and conversely, a glycine in the same position in CadC allows the protein to bind Zn(II) but not as a regulatory metal ion.
FIGURE 6.
Comparison of the structures of SmtB and CadC with and without bound Zn(II) in Site 2. A, in SmtB, Arg87 moves toward Site 2 upon Zn(II) binding. The backbone forms hydrogen bonds with the side chain His117. In addition, the side chain forms hydrogen bonds with the side chain of Glu120 and the backbone carboxyl group of His117. B, changes in the conformation of Arg87 influences the orientation of the first helix of the DNA binding site. C, the CadC residue corresponding to SmtB Arg87 is Gly84. D, interaction of Gly84 with His114 does not create any significant changes in the orientation of CadC helix 5. In each panel the structure in yellow is without zinc, and the structure in green is with bound zinc. The purple ball represents a zinc atom.
Because a wide variety of ArsR/SmtB family members use cysteine residues for metal binding, we also speculate that these sites were the ancestral regulatory sites and that the use of Site 2 for zinc regulation is a more recent adaptation. Alignment of the sequence of the proteins closely related to CadC and ArsR indicates that a glycine residue is highly conserved at the position corresponding to Arg87 of SmtB (supplemental Fig. 4). On the other hand, in SmtB homologues the most frequent residues at this position are basic: arginine, lysine, or histidine (23). It is interesting that only a single G-to-C substitution in the first position of all four glycine codons (GGN (where N represents any nucleotide)) is sufficient to convert them to arginine codons (CGN), which implies that a single point mutation might be sufficient to convert the non-regulatory Site 2 of a repressor ancestral to both CadC and SmtB to a regulatory function.
Supplementary Material
The atomic coordinates and structure factors (code 3F72) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants AI043428 (to B. P. R.) and DK068139 (to T. L. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–4.
Footnotes
The abbreviations used are: o/p, operator/promoter; XAS, x-ray absorption spectroscopy; EXAFS, extended x-ray fine structure; ICP-MS, inductively coupled plasma mass spectrometry; SSRL, Stanford Synchrotron Radiation Laboratory; NSLS, National Synchrotron Light Source; ITC, isothermal titration calorimetry; O/N, oxygen/nitrogen; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Mukhopadhyay, R., Rosen, B. P., Phung le, T., and Silver, S. (2002) FEMS Microbiol. Rev. 26 311-325 [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee, H., and Rosen, B. P. (2007) in Molecular Microbiology of Heavy Metals (Nies, D. H. S., and Silver, S., eds) pp. 371-406, Springer-Verlag New York Inc., New York
- 3.Corbisier, P., Ji, G., Nuyts, G., Mergeay, M., and Silver, S. (1993) FEMS Microbiol. Lett. 110 231-238 [DOI] [PubMed] [Google Scholar]
- 4.Busenlehner, L. S., Cosper, N. J., Scott, R. A., Rosen, B. P., Wong, M. D., and Giedroc, D. P. (2001) Biochemistry 40 4426-4436 [DOI] [PubMed] [Google Scholar]
- 5.Sun, Y., Wong, M. D., and Rosen, B. P. (2001) J. Biol. Chem. 276 14955-14960 [DOI] [PubMed] [Google Scholar]
- 6.Nucifora, G., Chu, L., Misra, T. K., and Silver, S. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 3544-3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong, M. D., Lin, Y. F., and Rosen, B. P. (2002) J. Biol. Chem. 277 40930-40936 [DOI] [PubMed] [Google Scholar]
- 8.Busenlehner, L. S., Weng, T. C., Penner-Hahn, J. E., and Giedroc, D. P. (2002) J. Mol. Biol. 319 685-701 [DOI] [PubMed] [Google Scholar]
- 9.Ye, J., Kandegedara, A., Martin, P., and Rosen, B. P. (2005) J. Bacteriol. 187 4214-4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner, J. S., Glands, P. D., Samson, A. C., and Robinson, N. J. (1996) Nucleic Acids Res. 24 3714-3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eicken, C., Pennella, M. A., Chen, X., Koshlap, K. M., VanZile, M. L., Sacchettini, J. C., and Giedroc, D. P. (2003) J. Mol. Biol. 333 683-695 [DOI] [PubMed] [Google Scholar]
- 12.Ellman, G. L. (1959) Arch. Biochem. Biophys. 82 70-77 [DOI] [PubMed] [Google Scholar]
- 13.Endo, G., and Silver, S. (1995) J. Bacteriol. 177 4437-4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzmic, P. (1996) Anal. Biochem. 237 260-273 [DOI] [PubMed] [Google Scholar]
- 15.Gill, S. C., and von Hippel, P. H. (1989) Anal. Biochem. 182 319-326 [DOI] [PubMed] [Google Scholar]
- 16.Wang, B., Alam, S. L., Meyer, H. H., Payne, M., Stemmler, T. L., Davis, D. R., and Sundquist, W. I. (2003) J. Biol. Chem. 278 20225-20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martell, A. E., and Smith, R. M. (1977) in Other Organic Ligands, p. 168, Plenum Press, New York
- 18.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 20.Ordonez, E., Thiyagarajan, S., Cook, J. D., Stemmler, T. L., Gil, J. A., Mateos, L. M., and Rosen, B. P. (2008) J. Biol. Chem. 283 25706-25714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichert, F., Bastolla, U., and Porto, M. (2007) BMC Bioinformatics 8 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook, W. J., Kar, S. R., Taylor, K. B., and Hall, L. M. (1998) J. Mol. Biol. 275 337-346 [DOI] [PubMed] [Google Scholar]
- 23.Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003) FEMS Microbiol. Rev. 27 131-143 [DOI] [PubMed] [Google Scholar]
- 24.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storonia, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.