Abstract
In archaea and eukarya, box C/D ribonucleoprotein (RNP) complexes are responsible for 2′-O-methylation of tRNAs and rRNAs. The archaeal box C/D small RNP complex requires a small RNA component (sRNA) possessing Watson-Crick complementarity to the target RNA along with three proteins: L7Ae, Nop5p, and fibrillarin. Transfer of a methyl group from S-adenosylmethionine to the target RNA is performed by fibrillarin, which by itself has no affinity for the sRNA-target duplex. Instead, it is targeted to the site of methylation through association with Nop5p, which in turn binds to the L7Ae-sRNA complex. To understand how Nop5p serves as a bridge between the targeting and catalytic functions of the box C/D small RNP complex, we have employed alanine scanning to evaluate the interaction between the Pyrococcus horikoshii Nop5p domain and an L7Ae box C/D RNA complex. From these data, we were able to construct an isolated RNA-binding domain (Nop-RBD) that folds correctly as demonstrated by x-ray crystallography and binds to the L7Ae box C/D RNA complex with near wild type affinity. These data demonstrate that the Nop-RBD is an autonomously folding and functional module important for protein assembly in a number of complexes centered on the L7Ae-kinkturn RNP.
Many biological RNAs require extensive modification to attain full functionality in the cell (1). Currently there are over 100 known RNA modification types ranging from small functional group substitutions to the addition of large multi-cyclic ring structures (2). Transfer RNA, one of many functional RNAs targeted for modification (3-6), possesses the greatest modification type diversity, many of which are important for proper biological function (7). Ribosomal RNA, on the other hand, contains predominantly two types of modified nucleotides: pseudouridine and 2′-O-methylribose (8). The crystal structures of the ribosome suggest that these modifications are important for proper folding (9, 10) and structural stabilization (11) in vivo as evidenced by their strong tendency to localize to regions associated with function (8, 12, 13). These roles have been verified biochemically in a number of cases (14), whereas newly emerging functional modifications are continually being investigated.
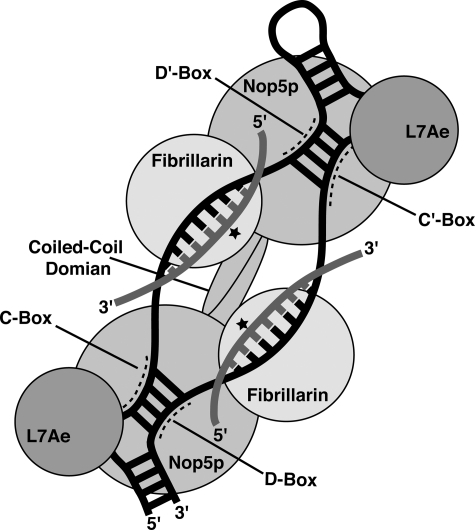
Box C/D ribonucleoprotein (RNP)3 complexes serve as RNA-guided site-specific 2′-O-methyltransferases in both archaea and eukaryotes (15, 16) where they are referred to as small RNP complexes and small nucleolar RNPs, respectively. Target RNA pairs with the sRNA guide sequence and is methylated at the 2′-hydroxyl group of the nucleotide five bases upstream of either the D or D′ box motif of the sRNA (Fig. 1, star) (17, 18). In archaea, the internal C′ and D′ motifs generally conform to a box C/D consensus sequence (19), and each sRNA contains two guide regions ∼12 nucleotides in length (20). The bipartite architecture of the RNP potentially enables the complex to methylate two distinct RNA targets (21) and has been shown to be essential for site-specific methylation (22).
FIGURE 1.
Organization of the archaeal box C/D complex. The protein components of this RNP are L7Ae, Nop5p, and fibrillarin, which together bind a box C/D sRNA. The regions of the Box C/D sRNA corresponding to the conserved C, D, C′, and D′ boxes are labeled. The target RNA binds the sRNA through Watson-Crick pairing and is methylated by fibrillarin at the fifth nucleotide from the D/D′ boxes (star).
In addition to the sRNA, the archaeal box C/D complex requires three proteins for activity (23): the ribosomal protein L7Ae (24, 25), fibrillarin, and the Nop56/Nop58 homolog Nop5p (Fig. 1). L7Ae binds to both box C/D and the C′/D′ motifs (26), which respectively comprise kink-turn (27) or k-loop structures (28), to initiate the assembly of the RNP (29, 30). Fibrillarin performs the methyl group transfer from the cofactor S-adenosylmethionine to the target RNA (31-33). For this to occur, the active site of fibrillarin must be positioned precisely over the specific 2′-hydroxyl group to be methylated. Although fibrillarin methylates this functional group in the context of a Watson-Crick base-paired helix (guide/target), it has little to no binding affinity for double-stranded RNA or for the L7Ae-sRNA complex (22, 26, 33, 34). Nop5p serves as an intermediary protein bringing fibrillarin to the complex through its association with both the L7Ae-sRNA complex and fibrillarin (22). Along with its role as an intermediary between fibrillarin and the L7Ae-sRNA complex, Nop5p possesses other functions not yet fully understood. For example, Nop5p self-dimerizes through a coiled-coil domain (35) that in most archaea and eukaryotic homologs includes a small insertion sequence of unknown function (36, 37). However, dimerization and fibrillarin binding have been shown to be mutually exclusive in Methanocaldococcus jannaschii Nop5p, potentially because of the presence of this insertion sequence (36). Thus, whether Nop5p is a monomer or a dimer in the active RNP is still under debate.
In this study, we focus our attention on the Nop5p protein to investigate its interaction with a L7Ae box C/D RNA complex because both the fibrillarin-Nop5p and the L7Ae box C/D RNA interfaces are known from crystal structures (29, 35, 38). Individual residues on the surface of a monomeric form of Nop5p (referred to as mNop5p) (22) were mutated to alanine, and the effect on binding affinity for a L7Ae box C/D motif RNA complex was assessed through the use of electrophoretic mobility shift assays. These data reveal that residues important for binding cluster within the highly conserved NOP domain (39, 40). To demonstrate that this domain is solely responsible for the affinity of Nop5p for the preassembled L7Ae box C/D RNA complex, we expressed and purified it in isolation from the full Nop5p protein. The isolated Nop-RBD domain binds to the L7Ae box C/D RNA complex with nearly wild type affinity, demonstrating that the Nop-RBD is truly an autonomously folding and functional module. Comparison of our data with the crystal structure of the homologous spliceosomal hPrp31-15.5K protein-U4 snRNA complex (41) suggests the adoption of a similar mode of binding, further supporting a crucial role for the NOP domain in RNP complex assembly.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis, Expression, and Purification—Pyrococcus horikoshii aFib (GI 14590005) and L7Ae (GI 74407908) were cloned, expressed, and purified essentially as previously described (22). Small changes in the protocol for L7Ae purification were made to improve the removal of contaminating nucleic acids. The supernatant was incubated at 80 °C for 30 min and centrifuged, and the resulting supernatant was adjusted to contain 6 m guanidinium hydrochloride, 25 mm sodium phosphate (pH 8.0), and 250 mm sodium chloride. This solution was loaded onto a nickel-nitrilotriacetic acid-agarose (Qiagen) column equilibrated in the same buffer. The protein was refolded on the column by washing the column with five column volumes of 50 mm Tris-Cl (pH 7.5), 300 mm sodium chloride, 10% glycerol, 5 mm β-mercaptoethanol, and 5 mm imidazole. L7Ae was eluted with five column volumes of 50 mm Tris-Cl (pH 7.5), 300 mm sodium chloride, 20 mm Na-EDTA, 10% glycerol, 5 mm β-mercaptoethanol, and 400 mm imidazole. L7Ae-containing fractions were subjected to TEV protease cleavage overnight to remove the hexahistidine tag followed by a second round of purification using nickel affinity chromatography.
P. horikoshii Nop5p (GI 14590006) and the mNop5p construct that contains a deletion of the coiled-coil were cloned, expressed, and purified as previously described (22). Alanine scanning was performed with the mNop5p protein previously shown to bind the L7Ae box C/D RNA complex with near wild type affinity (22) and has improved solution behavior. mNop5p alanine mutations were produced using site-directed PCR mutagenesis with the QuikChange site-directed mutagenesis kit (Stratagene) using a staggered primer design to avoid the occurrence of primer dimers (42). The conserved NOP domain construct (referred to as Nop-RBD) consists of residues 119-390 of P. horikoshii Nop5p, with the coiled-coil domain (residues 139-242) replaced with a Gly-Ala-Gly-Gly linker to eliminate the possibility of dimerization. This protein was cloned into a pET30 vector such that it is fused to an N-terminal hexahistidine-tagged maltose-binding protein with a TEV protease cleavage site between the fusion and NOP proteins. TEV cleavage results in a single N-terminal glycine residue not present in the native sequence. Nop-RBD was purified using a protocol identical to that for mNop5p and Nop5p.
RNA Synthesis and Labeling—RNA was transcribed, purified, and labeled with fluorescein as previously described (22). The sequence of the box C/D motif RNA used in binding experiments is: 5′-GGCACUGACCUCGAAAGAGGAAUGAUGAUU-3′.
Electrophoretic Mobility Shift Assays—Assays to measure the apparent equilibrium dissociation constant (KD) of mNop5p alanine mutants and the Nop-RBD for the L7Ae box C/D RNA complex were performed with 12% acrylamide gels (acrylamide:bis-acrylamide, 29:1) as described previously (22). Binding reactions for both mNop5p and Nop-RBD were conducted at 25 °C in a buffer containing 16 mm K+-HEPES, pH 7.5, 80 mm KCl, 0.01% Nonidet P-40, 25 μg/ml tRNA, and 2 nm labeled RNA. All of the gel shifts were performed in triplicate, and associated error was reported as one standard deviation from the mean. All of the alanine mutants of Nop5p reported in this work yielded an electrophoretic gel shift in the presence of the L7Ae box C/D RNA complex consistent with that of the wild type protein, indicating that each mutant was properly folded. Also, in calculating the KD for each mutant, we assumed that each protein was 100% active.
Crystallization and Data Collection—Crystals of Nop-RBD were grown using the hanging drop vapor diffusion method. Crystallization was achieved by adding 2 μl of a 14 mg/ml solution of Nop-RBD in 10 mm Na-MES, pH 6.0, to 2 μl of mother liquor containing 500 mm potassium iodide, 2% polyethylene glycol 3350 and incubating at 17 °C. Diffraction quality crystals grew to maximal size (∼400 μm in each dimension) within a week. The crystals were cryoprotected in mother liquor containing 30% ethylene glycol for at most 10 min and flash frozen in liquid nitrogen. A 360° data set was collected using Cu-Kα x-ray radiation using an R-AXIS IV++ detector on RU-200/Confocal blue optic source (Rigaku MSC).
Data Processing and Refinement of Structure—The diffraction images were indexed and integrated using MOSFLM and SCALA as part of the CCP4 suite (43). Analysis of the self-rotation function with MOLREP showed peaks at 0, 45, and 90° for χ = 180° and a single peak at 0.0° at χ = 90°, suggesting a P422 point group. Analysis of the integrated intensities with POINTLESS indicated that the space group was either P43212 or its enantiomer P41212. The intensities were then merged to the P422 point group using SCALA. PHENIX (44) XTRIAGE was used to analyze the intensity statistics. Anomalous signal from the iodide was found to 3.2 Å, and there was no evidence significant pseudotranslation via the native translational Patterson function or twinning via the multivariate Z score L test. Because anomalous signal was found in the data set, PHE-NIX AUTOSOL was used to determine the heavy atom substructure and calculate single-wavelength anomalous dispersion experimental phases. Six refined iodide sites were found with a figure of merit of 0.28 and 0.48 (pre- and post-statistical density modification, respectively). Inspection of the experimental density map showed clear features corresponding to Nop-RBD secondary structure, and the correct enantiomorphic space group of P41212 was determined from the maps. Initial model building using PHENIX AUTO-BUILD resulted in a model containing 50 sequence assigned residues and Rwork and Rfree values of 44 and 47%, respectively. Iterative model building in COOT (45) and phased maximum likelihood refinement in PHENIX resulted in a final model containing 115 of the 169 residues included in the construct and final Rwork and Rfree values of 31 and 34% at 2.5-Å resolution. Despite no additional unmodeled density in the |2Fo| - |Fc| maps greater than 2 σ, the R factors remained unacceptably high for this resolution. The data were then expanded into the P41 space group. Intensity statistics analysis by the Yeates Twinning server (46) suggested perfect merohedral twinning. The model built in the P422 point group was considered for molecular replacement generating two molecules in the asymmetric unit related by non-crystallographic symmetry. Manual model editing in COOT and coordinate, group atomic displacement factor, and twin fraction refinement with PHENIX lead to a final model with an Rwork value of 25.2%, an Rfree value of 28.6%, and a refined twinning fraction of 50%. Residues at both the N and C termini and the 15-amino acid linker region remained disordered in both the experimental and final model maps. Analysis of the final model in PROCHECK (47) showed 85.5% of the residues in the favored region, 13.5% of residues in the allowed region, and 1.0% of the residues in the outlier region. The model and structure factors have been deposited in the RCSB Protein Data Bank (3GQU and 3GQX).
RESULTS
Mapping the Binding Surface of Nop5p by Mutational Analysis Reveals a Distinct Region Responsible for Binding to the L7Ae Box C/D RNA Complex—To identify the region of the surface of Nop5p responsible for high affinity interaction with the L7Ae box C/D complex, we employed a comprehensive mutational analysis approach in which likely surface residues were converted to alanine and changes in binding affinity were determined. The sequence and crystal structure of the Archaeoglobus fulgidus Nop5p-fibrillarin dimer (35) were used to determine which amino acids most likely reside on the surface by sequence alignment with our P. horikoshii variant. Alanine scanning was performed with the mNop5p construct containing an internal deletion that prevents its self-dimerization but still allows for it to bind the L7Ae box C/D RNA complex with near wild type affinity (22). The apparent equilibrium dissociation constant (KD) for each mutant was determined by titrating the preformed L7Ae box C/D RNA complex with the mNop5p-fibrillarin heterodimer and quantifying the binding using an electrophoretic mobility shift assay.
Our mutational strategy involved changing conserved charged residues to alanine to find residues that potentially interact with the negatively charged phosphate backbone of the box C/D RNA. Residues were initially chosen to maximize surface coverage, but because the region responsible for binding to the L7Ae box C/D RNA complex emerged, our mutations became increasingly biased toward this part of the surface of the protein. In total, 31 mutations (Table 1) were made including two residues (Arg299 and Lys306) that resided within a 20-amino acid region disordered in the Nop5p-fibrillarin crystal structure (35). Neither of these residues contributed to L7Ae box C/D RNA complex binding. The region in direct contact with fibrillarin in the A. fulgidus structure was not explored except for one residue (Phe347), because the function of this portion of the surface of Nop5p has been clearly established (35).
TABLE 1.
Alanine scanning of mNop5p activity
| Mutant | KD | Krela | ΔΔ G | p value (t test)b |
|---|---|---|---|---|
| nm | kcal/mol | |||
| Wild typec | 76 ± 6 | 1 | ||
| K32A | 190 ± 31 | 2.5 | −0.54 | 0.0033 |
| E44A | 87 ± 37 | 1.1 | −0.06 | 0.6380 |
| K47A | 71 ± 17 | 0.93 | 0.04 | 0.6560 |
| E100A | 140 ± 17 | 1.8 | −0.35 | 0.0035 |
| E104A | 91 ± 15 | 1.2 | −0.11 | 0.1831 |
| F107A | 83 ± 12 | 1.1 | −0.06 | 0.4173 |
| N112A | 200 ± 62 | 2.6 | −0.57 | 0.0261 |
| K131A | 64 ± 8 | 0.84 | 0.10 | 0.1062 |
| D253A | 120 ± 9 | 1.6 | −0.28 | 0.0021 |
| K265A | 62 ± 11 | 0.81 | 0.12 | 0.1251 |
| E279A | 210 ± 41 | 2.7 | −0.59 | 0.0050 |
| S286A | 600 ± 60 | 7.9 | −1.22 | 0.0001 |
| T287A | 230 ± 6 | 3.0 | −0.65 | 0.0001 |
| Q289A | 1500 ± 81 | 20 | −1.77 | 0.0001 |
| R299A | 57 ± 11 | 0.75 | 0.17 | 0.0584 |
| K306A | 87 ± 20 | 1.1 | −0.06 | 0.4132 |
| N320A | 140 ± 3 | 1.8 | −0.35 | 0.0001 |
| W324A | 150 ± 13 | 2.0 | −0.41 | 0.0009 |
| Q326A | 280 ± 9 | 3.7 | −0.78 | 0.0001 |
| R327A | 50 ± 6 | 0.66 | 0.25 | 0.0061 |
| K329A | 210 ± 3 | 2.8 | −0.61 | 0.0001 |
| R332A | 1600 ± 240 | 21 | −1.80 | 0.0004 |
| K337A | 360 ± 23 | 4.7 | −0.92 | 0.0001 |
| F347A | 760 ± 100 | 10 | −1.36 | 0.0003 |
| E350A | 150 ± 12 | 2.0 | −0.41 | 0.0007 |
| E354A | 140 ± 24 | 1.8 | −0.35 | 0.0110 |
| E361A | 110 ± 23 | 1.4 | −0.20 | 0.0684 |
| R363A | 400 ± 34 | 5.3 | −0.99 | 0.0001 |
| K365A | 130 ± 15 | 1.7 | −0.31 | 0.0044 |
| E366A | 260 ± 37 | 3.4 | −0.72 | 0.0010 |
| K368A | 130 ± 3 | 1.7 | −0.31 | 0.0002 |
Krel = KD,mutant/KD,wild type.
p values from a two-tailed unpaired t test. The mutations to mNop5p that result in a statistically significant change in binding affinity relative to wild type are listed in bold type (95% confidence level).
Wild type refers to the mNop5p protein that contains a deletion of the coiled-coil domain of Nop5, preventing its dimerization.
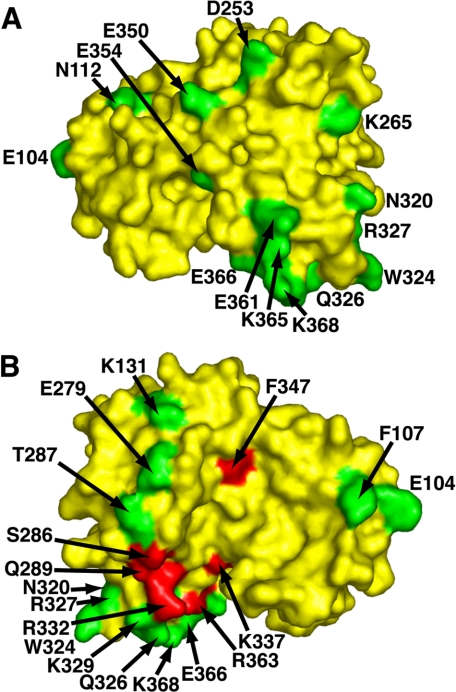
The mutational analysis revealed that the L7Ae box C/D RNA-binding surface is localized to a limited region of Nop5p (Fig. 2). One face of Nop5p does not contribute to the L7Ae box C/D RNA interaction, as evidenced by the observation that all alanine mutations associated with this region of the protein have no more than a 2-3-fold affect on binding affinity (Lys32, Glu44, Lys47, Glu100, Glu104, Asn112, Asp253, Lys265, Asn320, Arg327, Glu350, Glu354, Glu361, Lys365, and Lys368; Fig. 2A and Table 1). On the opposite face of Nop5p, there is a clearly defined region where alanine mutations produce a pronounced reduction in L7Ae box C/D RNA complex binding affinity (S286, T287, Q289, Q326, R332, K337, R363, F347, and E366; Fig. 2B and Table 1). Residues peripheral to this place on Nop5p have little affect on binding (Phe107, Lys131, Glu279, Trp324, and Lys329). A previous study identified a single residue (Arg224 in A. fulgidus Nop5p) that abolished binding upon mutation to alanine (35). The equivalent residue in P. horikoshii Nop5p (Arg332) is located in the region important for binding, and mutation of this residue results in a significant decrease in binding affinity, indicating that the two Nop5p variants behave similarly with respect to L7Ae box C/D RNA recognition.
FIGURE 2.
The residues in mNop5p important for L7Ae box C/D RNA binding localize to one region. The backside of Nop5p (A) is virtually devoid of residues important for the interaction. However, the front side (B) exhibits a well defined cluster of residues important for binding. Compared with wild type mNop5p binding affinity, alanine mutations that result in less than a 4-fold change are shown in green, and mutations resulting in a greater than 4-fold change in binding affinity are assigned to red. Residues with a greater than 2-fold change in binding affinity upon mutation to alanine are labeled. Several mutated residues listed in Table 1 are not visible in the A. fulgidus structure used for surface residue mapping. These residues either occur in disordered regions (Arg299 and Lys306) or lack an equivalent in the A. fulgidus structure (Lys32, Glu44, Lys47, and Glu100).
This analysis revealed that all of the residues important for L7Ae box C/D RNA binding reside within the NOP domain (39, 40) that has been previously implicated in L7Ae-sRNA (35) and 15.5K protein-U4 snRNA binding (48). These data suggest the possibility that the conserved NOP domain of Nop5p is a modular, independently functional domain in the absence of both the coiled-coil and the fibrillarin-binding regions. Therefore, we expressed a protein corresponding exclusively to this region (referred to as Nop-RBD) for further analysis.
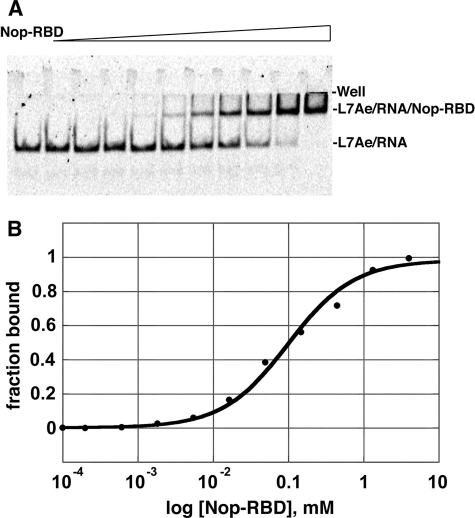
The NOP Domain of Nop5p Exhibits near Wild Type Affinity for the L7Ae Box C/D RNA Complex—Nop-RBD was purified by the same protocol used for mNop5p and full Nop5p (22). This protocol requires high salt concentrations (1 m NaCl) to enhance the solubility of the mNop5p and full Nop5p proteins. Moreover, mNop5p requires very high salt concentrations to achieve activity in the context of the box C/D RNP (22). Removal of the fibrillarin-binding domain of mNop5p to create Nop-RBD alleviated this requirement for high salt resulting in a stable, well behaved protein under low ionic strength conditions. The NOP domain binds to the L7Ae box C/D motif RNA complex with nearly identical affinity as mNop5p (Fig. 3); mNop5p binds the complex with an apparent KD of 76 ± 6 nm, whereas the NOP domain binds with an apparent KD of 106 ± 4 nm. These data clearly show that residues in the fibrillarin-binding region of Nop5p (residues 1-118) are not contributing to the L7Ae box C/D RNA interaction. As expected, Nop-RBD has no detectable affinity for fibrillarin, as demonstrated through a gel mobility shift assay where fibrillarin is titrated against the L7Ae-Nop-RBD box C/D RNA ternary complex (data not shown).
FIGURE 3.
L7Ae-RNA complex binding to the Nop-RBD. A, formation of the ternary complex was monitored using an electrophoretic mobility shift assay. The far left lane was without Nop-RBD, and the subsequent lanes had increasing amounts of protein in concentrations ranging from 200 pm to 4 μm. The fluorescein-labeled L7Ae box C/D RNA complex was at 15 nm. B, the resulting data from a single experiment fit to a standard single-site binding model (KD averaged over three experiments = 106 ± 4 nm).
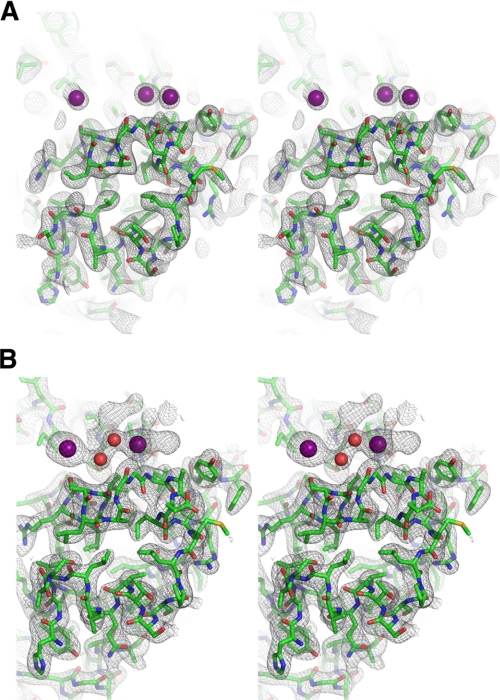
The Conserved NOP Domain Is an Independently Folded Domain—The improvement in solution behavior of the P. horikoshii Nop5p NOP domain makes it an excellent target for crystallization both in isolation and in a variety of subcomplexes of the box C/D RNP complex. To validate this protein as a crystallization target, we crystallized and solved the structure of the Nop-RBD. We were able to obtain crystals that diffracted x-ray radiation to ∼2.5-Å resolution and collected data of sufficient quality to yield a model of the NOP domain (Figs. 4 and 5A and Table 2). Notably, several regions of the protein including the first 25 residues of the construct (consisting of an N-terminal glycine, residues 119-138, and the Gly-Gly-Ala-Gly linker replacing the coiled-coil domain), an internal region consisting of 16 amino acids (294-309), and the 17 C-terminal residues (374-390) were not observed in the resulting electron density map. Thus, approximately one-third of the protein mass could not be accounted for by the model, directly resulting in higher final Rwork and Rfree values for a structure at this resolution.
FIGURE 4.
A, stereoview of the experimental density map in the P41212 space group contoured at 1 σ. The iodides are represented as purple spheres. B, stereoview of the |2Fo| - |Fc| map in the P41 space group, contoured at 1 σ. The iodides are represented as purple spheres, and waters are red spheres.
FIGURE 5.
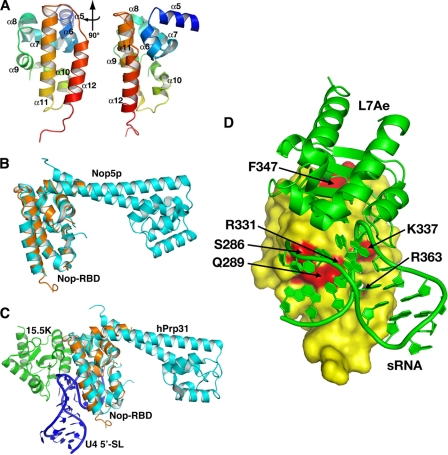
A, ribbon diagram of the crystal structure of the isolated Nop-RBD solved to 2. 5 Å resolution. The protein chain was colored as a gradient from blue (N terminus, residue 243) to red (C terminus, residue 373), and the α-helices were labeled using the nomenclature for the intact Nop5p protein (35). Two perspectives are shown with the right representing a 90° rotation of the structure on the left. B, the isolated Nop-RBD structure superimposes upon the P. furiosus Nop5p-fibrillarin structure (Protein Data Bank code 2NNW) with an RMSD of 0.77 Å over all amino acids common between the two structures. Fibrillarin and the fibrillarin-binding domain of Nop5p have been omitted for clarity. C, the isolated Nop-RBD structure superimposes upon the hPrp31 structure (Protein Data Bank code 2OZB) with an RMSD of 1.6 Å over all amino acids common between the two structures. Prp31 is shown in complex with 15.5K protein (an L7Ae homolog) and the 5′-stem-loop of the kink-turn containing U4 snRNA. D, model of the Nop-RBD interaction with the L7Ae box C/D RNA complex (Protein Data Bank code 1RLG) (35) using the hPrp31-15.5K-U4 snRNA complex (Protein Data Bank code 2OZB) (41) as a guide for orienting the box C/D components. L7Ae box C/D RNA complex (green) is docked against a surface representation of the Nop-RBD (yellow). Residues in mNop5p whose mutation to alanine strongly affects binding (Krel > 4) are highlighted in red, underscoring the consistency between the biochemical and structural data.
TABLE 2.
Crystallographic data and refinement statistics
| Data collection | |
| Space group | P41212 |
| Unit cell parameters | |
| a, b, c (Å) | 73.6, 73.6, 114.3 |
| α, β, γ | 90°, 90°, 90° |
| Resolution (Å) | 25−2.5 (2.76−2.50)a |
| Observed reflections | 433,471 |
| Unique reflections | 19,908 |
| Completeness (%) | 99.9 (100.0) |
| Rp.i.m.b | 0.106 (0.389) |
| <I/σ> | 46.5 (2.8) |
| Redundancy | 27.3 (28.1) |
| Experimental phasing | |
| Space group | P41212 |
| No. of iodide sites | 6 |
| Figure of merit (before/after density modification) | 0.284/0.480 |
| Refinement | |
| Space group | P41 |
| Twinning (%) | 50.0 |
| Twinning operator | h, −k, −l |
| Resolution (Å) | 23.6−2.5 (2.63−2.50) |
| Reflections used | 19,699 |
| Rwork (%)c | 25.2 (33.9) |
| Rfree (%) | 28.6 (33.7) |
| Model quality | |
| RMSD bond length (Å) | 0.004 |
| RMSD bond angles | 0.865° |
| Luzzati coordinate error (Å) | 0.523 |
Numbers in parentheses correpond to the highest resolution shell.
Rp.i.m. = ∑hkl[1/(N − 1)]½∑i|Ii(hkl) − hkl|/∑hkl∑iIi (hkl).
R = 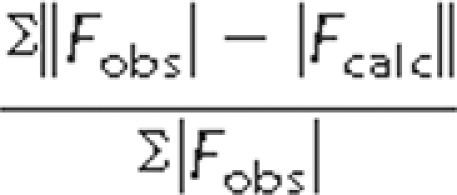 .
.
The resulting model when superimposed with the crystal structures of the full Nop5p protein from A. fulgidus (35) (Protein Data Bank code 1NT2) and P. furiosus (49) (Protein Data Bank code 2NNW) reveals an identical fold with RMSDs of 1.06 and 0.77 Å, respectively (Fig. 5B). Likewise, the P. horikoshii Nop-RBD superimposes well with the homologous region of the human Prp31 protein of the spliceosome (Protein Data Bank code 2OZB) with an RMSD of 1.61 Å (Fig. 5C). This structure thus reinforces the idea that the NOP domain is an independently folding and active RNP binding module (41).
DISCUSSION
In many RNP enzymes, there is a division of labor between the protein and RNA components. In box C/D, box H/ACA, telomerase, and the RNA-induced silencing complex, RNA serves to direct the enzyme to the appropriate target and thus dictates the specificity of the enzyme (37, 50, 51). On the other hand, the protein components of each of these enzymes perform the catalytic role by acting on the guide sRNA-target complex; in the box C/D RNPs this is the role of the S-adenosylmethionine-dependent methyltransferase fibrillarin (32-34). Thus, a central feature of these enzymes is the coordination of targeting and specificity determination by RNA and catalysis by proteins to achieve both high specificity and activity in the cell. In the case of the box C/D RNPs, this role appears to be played by the conserved protein Nop5p in archaea and the Nop56/58 proteins in eukaryotes (35, 37, 52, 53).
Nop5p serves as the intermediary between specificity and catalytic activity by virtue of its ability to bind to both fibrillarin and the L7Ae box C/D RNP complex. Its interaction with the L7Ae-sRNA RNP is particularly notable as the kink-turn motif, formed by the box C/D sequence motif, is a fundamental building block of tertiary architecture within many biological RNAs (24, 25, 38, 54). Despite its essential role in the function of the box C/D RNP (55-57), the basic details of how Nop5p serves as an intermediary between the box C/D motif of the sRNA and the active site of fibrillarin have only just begun to be revealed. In this study we have sought to improve our understanding of this interaction by an extensive mutational analysis aimed at identifying the region of Nop5p necessary for interaction with the L7Ae box C/D region of the archaeal box C/D small RNP complex.
The region of Nop5p important for binding to the L7Ae box C/D RNA complex has been suspected to occur within a highly conserved region called the NOP conserved domain that exists in other RNP complexes (35, 40, 48, 55, 58). These complexes exhibit diverse functionality but have in common an L7Ae homolog-kink-turn complex necessary for nucleation of complex assembly. In the spliceosome, for example, the conserved region of Nop5p is present in the hPrp31 (48) and 61K (58) proteins that recognize the 15.5K protein-U4 snRNA complex. Evidence for assigning an L7Ae box C/D RNA binding function to the conserved region of Nop5p is the presence of a large patch of surface residues whose identities are important for maintaining the affinity of Nop5p binding to the L7Ae box C/D RNP complex. Mutation of a single residue in this region (Arg332 in this study) has a strongly deleterious effect on the ability of the Nop5p protein to interact with the L7Ae-kink-turn complex (35). Furthermore, in the spliceosome, a peptide within this conserved region cross-links to the kink-turn RNA, demonstrating direct interaction between the 61K protein and the 15.5K protein-U4 snRNA complex (48).
Our mutational analysis of the surface of Nop5p revealed that all residues shown to be important for high affinity and specific binding reside within the conserved NOP domain. Expression of the isolated NOP domain (Nop-RBD) demonstrates that it is capable of interacting with the L7Ae box C/D RNA complex with nearly the same affinity as the full protein. Thus, the conserved NOP domain represents a structurally distinct RNP-binding protein. Additionally, the crystal structure of Nop-RBD is extremely similar to the NOP domains of full-length Nop5p and hPrp31, indicating that this is a bona fide RNP-binding domain. Submission of the structure of this domain to VAST (59, 60) and DALI (61, 62) to find potential structural neighbors results in both Nop5p and hPrp31 being returned but no other significant hits. Thus, the conserved NOP domain represents a structurally distinct RNP-binding protein.
A crystal structure of a ternary spliceosomal subcomplex composed of hPrp31 (Nop5p homolog), the 15.5K protein (L7Ae homolog), and a kink-turn containing fragment of the U4 snRNA (Protein Data Bank code 2OZB) (41) has been solved. The conserved NOP domain of hPrp31 shares considerable sequence identity with the box C/D Nop5p protein examined in this study (Fig. 6). Superposition of the Nop-RBD structure solved in this study with the ternary spliceosomal RNP via the conserved NOP domain yields a very reasonable hypothetical model of the ternary interaction in the archaeal box C/D rRNP (Fig. 5D). Residues found important for the binding of Nop5p to the box C/D RNA complex correlate well with residues in hPrp31 directly in contact with the U4 snRNA. The precise details of the interaction, specifically how Nop5p discriminates between the U4 stem II and the box C/D stem, assuming an overall similar interface through the conserved NOP RNA-binding domain, must await further structural studies. Nevertheless, our data clearly support a composite protein-protein and protein-RNA interface similar to that observed in the homologous complex of the U4 snRNP (58).
FIGURE 6.
Nop-RBD sequence alignment. The isolated Nop-RBD domain of P. horikoshii is aligned with the homologous regions of A. fulgidus Nop5p (Afu_Nop5p), M. jannaschii Nop5p (Mja_Nop5p), Saccharomyces cerevisiae Nop58 (Sce_Nop58), Drosophila melanogaster Nop56 (Dme_Nop56), Homo sapiens Nop58 (Has_Nop58), S. cerevisiae Prp31 (Sce_Prp31), and H. sapiens Prp31 (Has_Prp31). Secondary structural elements apparent in the Nop-RBD crystal structure are annotated above the sequence and numbered according to the nomenclature for the intact Nop5p protein (35). The open bars represent regions disordered in either the Nop-RBD structure (above sequence) or the hPrp structure (below sequence). The cyan bars indicate regions of the protein not present in the crystallized construct. These are the coiled-coil domain of Nop5p (above sequence) and the C-terminal region of hPrp31 (below sequence). The circles indicate residues mutated to alanine in this study and are colored in the same manner as for Fig. 2. Compared with wild type mNop5p binding affinity, alanine mutations that result in less than a 4-fold change are shown in green, and mutations resulting in a greater than 4-fold change in binding affinity are assigned to red. The squares indicate residues in hPrp31 predicted to interact with 15.5K protein (yellow), U4 RNA (green), or both (red) (41) based upon the crystal structure (Protein Data Bank code 2OZB).
Acknowledgments
We thank Johnny Croy and members of the Batey laboratory for critically reading this manuscript.
The atomic coordinates and structure factors (codes 3GQU and 3GQX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant GM08953 (to R. T. B.). This work was also supported by a graduate research training grant (to F. E. R.) through the University of Colorado, Boulder.
Footnotes
The abbreviations used are: RNP, ribonucleoprotein; sRNA, small RNA; RMSD, root mean square deviation; MES, 4-morpholineethanesulfonic acid; snRNA, small nucleolar RNA.
References
- 1.Grosjean, H., and Benne, R. (1998) Modification and Editing of RNA, American Society for Microbiology Press, Washington, DC
- 2.Rozenski, J., Crain, P. F., and McCloskey, J. A. (1999) Nucleic Acids Res. 27 196-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clouet-d'Orval, B., Gaspin, C., and Mougin, A. (2005) Biochimie (Paris) 87 889-895 [DOI] [PubMed] [Google Scholar]
- 4.Decatur, W. A., and Fournier, M. J. (2003) J. Biol. Chem. 278 695-698 [DOI] [PubMed] [Google Scholar]
- 5.Kiss, T. (2001) EMBO J. 20 3617-3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziesche, S. M., Omer, A. D., and Dennis, P. P. (2004) Mol. Microbiol. 54 980-993 [DOI] [PubMed] [Google Scholar]
- 7.Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. (1998) Nucleic Acids Res. 26 148-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decatur, W. A., and Fournier, M. J. (2002) Trends Biochem. Sci. 27 344-351 [DOI] [PubMed] [Google Scholar]
- 9.Sirum-Connolly, K., and Mason, T. L. (1993) Science 262 1886-1889 [DOI] [PubMed] [Google Scholar]
- 10.Sirum-Connolly, K., and Mason, T. L. (1995) Nucleic Acids Symp. Ser. 73-75 [PubMed]
- 11.Cunningham, P. R., Richard, R. B., Weitzmann, C. J., Nurse, K., and Ofengand, J. (1991) Biochimie (Paris) 73 789-796 [DOI] [PubMed] [Google Scholar]
- 12.Peculis, B. A. (1997) Mol. Cell. Biol. 17 3702-3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tycowski, K. T., Shu, M. D., and Steitz, J. A. (1994) Science 266 1558-1561 [DOI] [PubMed] [Google Scholar]
- 14.Helm, M. (2006) Nucleic Acids Res. 34 721-733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis, P. P., Omer, A., and Lowe, T. (2001) Mol. Microbiol. 40 509-519 [DOI] [PubMed] [Google Scholar]
- 16.Omer, A. D., Lowe, T. M., Russell, A. G., Ebhardt, H., Eddy, S. R., and Dennis, P. P. (2000) Science 288 517-522 [DOI] [PubMed] [Google Scholar]
- 17.Kiss-Laszlo, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M., and Kiss, T. (1996) Cell 85 1077-1088 [DOI] [PubMed] [Google Scholar]
- 18.Tycowski, K. T., Smith, C. M., Shu, M. D., and Steitz, J. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14480-14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaspin, C., Cavaille, J., Erauso, G., and Bachellerie, J. P. (2000) J. Mol. Biol. 297 895-906 [DOI] [PubMed] [Google Scholar]
- 20.Tran, E., Zhang, X., Lackey, L., and Maxwell, E. S. (2005) RNA 11 285-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, C. M., and Steitz, J. A. (1997) Cell 89 669-672 [DOI] [PubMed] [Google Scholar]
- 22.Hardin, J. W., and Batey, R. T. (2006) Nucleic Acids Res. 34 5039-5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omer, A. D., Ziesche, S., Decatur, W. A., Fournier, M. J., and Dennis, P. P. (2003) Mol. Microbiol. 48 617-629 [DOI] [PubMed] [Google Scholar]
- 24.Kuhn, J. F., Tran, E. J., and Maxwell, E. S. (2002) Nucleic Acids Res. 30 931-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozhdestvensky, T. S., Tang, T. H., Tchirkova, I. V., Brosius, J., Bachellerie, J. P., and Huttenhofer, A. (2003) Nucleic Acids Res. 31 869-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran, E. J., Zhang, X., and Maxwell, E. S. (2003) EMBO J. 22 3930-3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szewczak, L. B., Gabrielsen, J. S., Degregorio, S. J., Strobel, S. A., and Steitz, J. A. (2005) RNA 11 1407-1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolivos, S., Carpousis, A. J., and Clouet-d'Orval, B. (2005) Nucleic Acids Res. 33 6507-6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, T., Zhang, Y., Fenley, M. O., and Li, H. (2004) Structure 12 807-818 [DOI] [PubMed] [Google Scholar]
- 30.Suryadi, J., Tran, E. J., Maxwell, E. S., and Brown, B. A., 2nd (2005) Biochemistry 44 9657-9672 [DOI] [PubMed] [Google Scholar]
- 31.Aittaleb, M., Visone, T., Fenley, M. O., and Li, H. (2004) J. Biol. Chem. 279 41822-41829 [DOI] [PubMed] [Google Scholar]
- 32.Deng, L., Starostina, N. G., Liu, Z. J., Rose, J. P., Terns, R. M., Terns, M. P., and Wang, B. C. (2004) Biochem. Biophys. Res. Commun. 315 726-732 [DOI] [PubMed] [Google Scholar]
- 33.Wang, H., Boisvert, D., Kim, K. K., Kim, R., and Kim, S. H. (2000) EMBO J. 19 317-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omer, A. D., Ziesche, S., Ebhardt, H., and Dennis, P. P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5289-5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aittaleb, M., Rashid, R., Chen, Q., Palmer, J. R., Daniels, C. J., and Li, H. (2003) Nat. Struct. Biol. 10 256-263 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, X., Champion, E. A., Tran, E. J., Brown, B. A., 2nd, Baserga, S. J., and Maxwell, E. S. (2006) RNA 12 1092-1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichow, S. L., Hamma, T., Ferre-D'Amare, A. R., and Varani, G. (2007) Nucleic Acids Res. 35 1452-1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamma, T., and Ferre-D'Amare, A. R. (2004) Structure 12 893-903 [DOI] [PubMed] [Google Scholar]
- 39.Makarova, O. V., Makarov, E. M., Liu, S., Vornlocher, H. P., and Luhrmann, R. (2002) EMBO J. 21 1148-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vithana, E. N., Abu-Safieh, L., Allen, M. J., Carey, A., Papaioannou, M., Chakarova, C., Al-Maghtheh, M., Ebenezer, N. D., Willis, C., Moore, A. T., Bird, A. C., Hunt, D. M., and Bhattacharya, S. S. (2001) Mol. Cell 8 375-381 [DOI] [PubMed] [Google Scholar]
- 41.Liu, S., Li, P., Dybkov, O., Nottrott, S., Hartmuth, K., Luhrmann, R., Carlomagno, T., and Wahl, M. C. (2007) Science 316 115-120 [DOI] [PubMed] [Google Scholar]
- 42.Zheng, L., Baumann, U., and Reymond, J. L. (2004) Nucleic Acids Res. 32 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project, N. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 44.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 45.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 46.Padilla, J. E., and Yeates, T. O. (2003) Acta Crystallogr. D. Biol. Crystallogr. 59 1124-1130 [DOI] [PubMed] [Google Scholar]
- 47.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 48.Nottrott, S., Urlaub, H., and Luhrmann, R. (2002) EMBO J. 21 5527-5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oruganti, S., Zhang, Y., Li, H., Robinson, H., Terns, M. P., Terns, R. M., Yang, W., and Li, H. (2007) J. Mol. Biol. 371 1141-1150 [DOI] [PubMed] [Google Scholar]
- 50.Mitchell, J. R., Cheng, J., and Collins, K. (1999) Mol. Cell. Biol. 19 567-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rana, T. M. (2007) Nat. Rev. Mol. Cell Biol. 8 23-36 [DOI] [PubMed] [Google Scholar]
- 52.Lafontaine, D. L., and Tollervey, D. (1999) RNA 5 455-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman, D. R., Kuhn, J. F., Shanab, G. M., and Maxwell, E. S. (2000) RNA 6 861-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000) Science 289 905-920 [DOI] [PubMed] [Google Scholar]
- 55.Gautier, T., Berges, T., Tollervey, D., and Hurt, E. (1997) Mol. Cell. Biol. 17 7088-7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verheggen, C., Mouaikel, J., Thiry, M., Blanchard, J. M., Tollervey, D., Bordonne, R., Lafontaine, D. L., and Bertrand, E. (2001) EMBO J. 20 5480-5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins, N. J., Dickmanns, A., and Luhrmann, R. (2002) Mol. Cell. Biol. 22 8342-8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz, A., Nottrott, S., Watkins, N. J., and Luhrmann, R. (2006) Mol. Cell. Biol. 26 5146-5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madej, T., Gibrat, J. F., and Bryant, S. H. (1995) Proteins 23 356-369 [DOI] [PubMed] [Google Scholar]
- 60.Gibrat, J. F., Madej, T., and Bryant, S. H. (1996) Curr. Opin. Struct. Biol. 6 377-385 [DOI] [PubMed] [Google Scholar]
- 61.Holm, L., and Sander, C. (1993) J. Mol. Biol. 233 123-138 [DOI] [PubMed] [Google Scholar]
- 62.Holm, L., and Sander, C. (1996) Methods Enzymol. 266 653-662 [DOI] [PubMed] [Google Scholar]