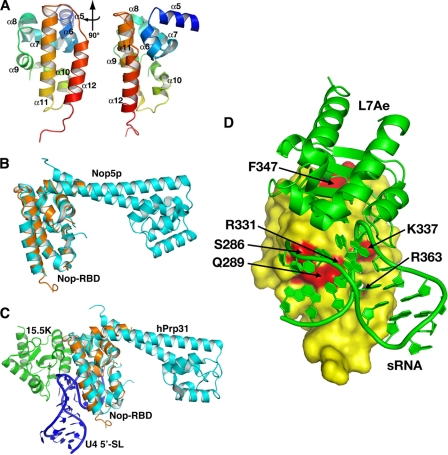
FIGURE 5.
A, ribbon diagram of the crystal structure of the isolated Nop-RBD solved to 2. 5 Å resolution. The protein chain was colored as a gradient from blue (N terminus, residue 243) to red (C terminus, residue 373), and the α-helices were labeled using the nomenclature for the intact Nop5p protein (35). Two perspectives are shown with the right representing a 90° rotation of the structure on the left. B, the isolated Nop-RBD structure superimposes upon the P. furiosus Nop5p-fibrillarin structure (Protein Data Bank code 2NNW) with an RMSD of 0.77 Å over all amino acids common between the two structures. Fibrillarin and the fibrillarin-binding domain of Nop5p have been omitted for clarity. C, the isolated Nop-RBD structure superimposes upon the hPrp31 structure (Protein Data Bank code 2OZB) with an RMSD of 1.6 Å over all amino acids common between the two structures. Prp31 is shown in complex with 15.5K protein (an L7Ae homolog) and the 5′-stem-loop of the kink-turn containing U4 snRNA. D, model of the Nop-RBD interaction with the L7Ae box C/D RNA complex (Protein Data Bank code 1RLG) (35) using the hPrp31-15.5K-U4 snRNA complex (Protein Data Bank code 2OZB) (41) as a guide for orienting the box C/D components. L7Ae box C/D RNA complex (green) is docked against a surface representation of the Nop-RBD (yellow). Residues in mNop5p whose mutation to alanine strongly affects binding (Krel > 4) are highlighted in red, underscoring the consistency between the biochemical and structural data.