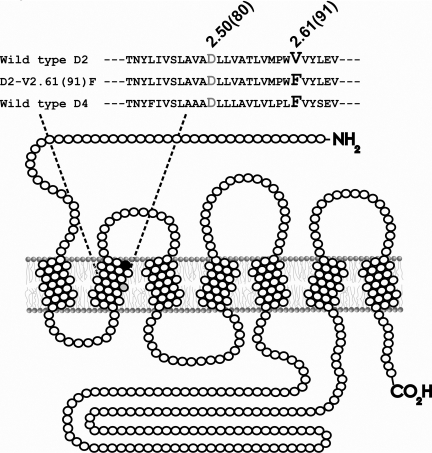
Fig. 1.
Depiction of the D2-V2.61(91)F mutant dopamine receptor as a
monomer in a section of lipid bilayer. This figure represents the unfolded
D2L receptor showing the amino terminus (-NH2) on the
extracellular side and the carboxyl terminus (-CO2H) on the
intracellular side. Open circles (○) are used to indicate wild-type amino
acids, whereas closed circles (• and
 ) are used to represent
specific amino acids. As shown in the sequence, a valine-to-phenylalanine
mutation at amino acid residue 91 (•) results in the
D2-V2.61(91)F mutant dopamine receptor. The purpose of the
V2.61(91)F mutation is to modify the binding pocket of the D2
receptor with the corresponding residue of the D4 receptor and make
it more accommodating to D4-selective 1,4-DAPs
(Simpson et al., 1999;
Schetz et al., 2000;
Kortagere et al., 2004;
Floresca et al., 2005).
Although most ligands bind an orthosteric binding site accessible from the
extracellular face of the receptor
(Floresca and Schetz, 2004),
the sodium ion binds the receptor through an intracellular allosteric binding
site formed by the interactions of transmembrane segments 2, 3, and 7
(Neve et al., 2001). Also
shown in the diagram and the sequence is the relative position of the
conserved negatively charged D2.50(80) that is critical for the interaction of
sodium ions with the dopamine receptor (Neve et al.,
1991,
2001).
) are used to represent
specific amino acids. As shown in the sequence, a valine-to-phenylalanine
mutation at amino acid residue 91 (•) results in the
D2-V2.61(91)F mutant dopamine receptor. The purpose of the
V2.61(91)F mutation is to modify the binding pocket of the D2
receptor with the corresponding residue of the D4 receptor and make
it more accommodating to D4-selective 1,4-DAPs
(Simpson et al., 1999;
Schetz et al., 2000;
Kortagere et al., 2004;
Floresca et al., 2005).
Although most ligands bind an orthosteric binding site accessible from the
extracellular face of the receptor
(Floresca and Schetz, 2004),
the sodium ion binds the receptor through an intracellular allosteric binding
site formed by the interactions of transmembrane segments 2, 3, and 7
(Neve et al., 2001). Also
shown in the diagram and the sequence is the relative position of the
conserved negatively charged D2.50(80) that is critical for the interaction of
sodium ions with the dopamine receptor (Neve et al.,
1991,
2001).