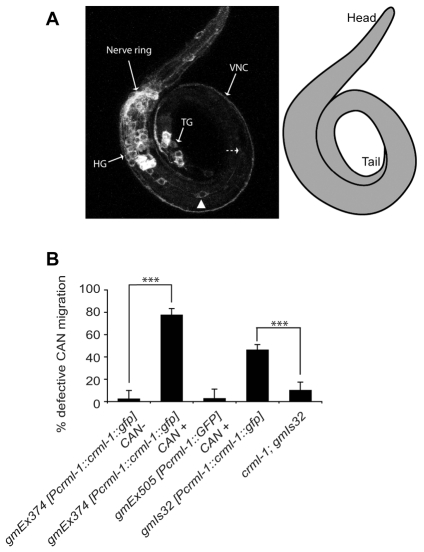
Fig. 3.
Excess CRML-1 disrupts CAN migration. (A) Confocal projection of an L1 C. elegans expressing the extrachromosomal array gmEx374 [Pcrml-1::crml-1::gfp]. CRML-1::GFP is expressed in neurons of the head (HG) and tail (TG) ganglia, and in axons that form the nerve ring, the ventral nerve cord (VNC) and the dorsal nerve cord (DNC) (not shown). The position of a CAN is indicated (arrowhead). This CAN is displaced anterior to its normal position, which is indicated by the dashed arrow. A schematic of the animal is presented on the right. (B) CAN migration defects in CRML-1 transgenic animals. See Fig. 2C legend for details. Extrachromosomal arrays are lost during mitosis, which allowed us to determine differences between animals expressing the array in CAN and those that had lost the array in CAN. For animals carrying gmEx374, we scored CANs in which we detected CRML-1::GFP in the cells (CAN+) and CANs in which we could not (CAN-). Statistically significant differences between CAN+ and CAN-animals and between gmIs32 and crml-1; gmIs32 are marked (***P<0.0001). For the distribution of CANs relative to P and V cells and the number of CANs scored, see Fig. S2 in the supplementary material. Error bars, s.e.m. At least 34 animals were scored per genotype.