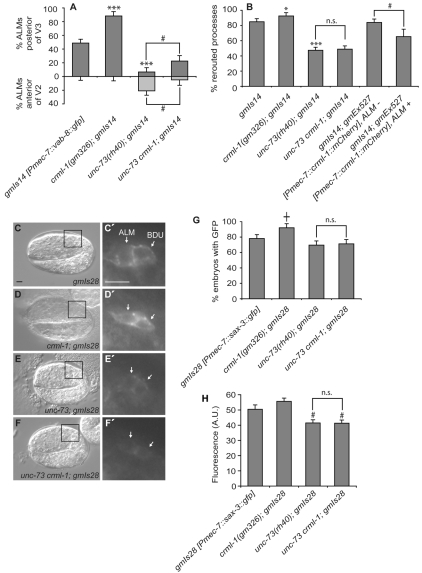
Fig. 6.
CRML-1 regulates VAB-8L-induced ALM abnormalities and SAX-3 levels. (A) Excessive ALM migration caused by VAB-8L misexpression is enhanced by loss of CRML-1 and suppressed by loss of UNC-73 Rac GEF activity. The percentage of ALMs found posterior of wild-type positions is depicted in dark gray (above x-axis), and the percentage of ALMs found anterior of wild-type positions is depicted in light gray (beneath x-axis). Statistically significant differences between gmIs14 [Pmec-7::vab-8::gfp] and crml-1; gmIs14 or unc-73; gmIs14 are marked (***P<0.0001). Differences between unc-73 crml-1; gmIs14 and unc-73; gmIs14 are indicated (#, P<0.03). unc-73 crml-1; gmIs14 was compared with unc-73; gmIs14. Error bars, s.e.m. For the distribution of ALMs relative to P and V cells and the number scored, see Fig. S1 in the supplementary material. At least 40 animals were scored per genotype. Animals were raised at 25°C. (B) VAB-8L-dependent ALM process rerouting is enhanced by loss of CRML-1 and suppressed by both the reduction of UNC-73 and overexpression of CRML-1. Statistically significant differences between gmIs14 and other strains are marked (***P<0.0001, *P<0.05); n.s., not significant. The statistically significant difference between gmIs14; gmEx527 strains bearing the array in ALM compared with those bearing the array but not in ALM is marked (#, P<0.03). At least 125 animals were scored per genotype, except gmIs14; gmEx527, ALM+ where n=26. (C-F′) Nomarski (C-F) and GFP (boxed regions enlarged in C′-F′) images of 2-fold C. elegans embryos expressing gmIs28 [Pmec-7::sax-3::gfp] in ALM and BDU neurons (arrows) just prior to cell migration. Anterior is to the left, dorsal is up. Scale bars: 5 μm. (G) Percentage of embryos in which a SAX-3::GFP signal was detected in ALM and BDU neurons during the 2-fold stage of embryogenesis. Strains were scored on the same day to ensure that the GFP signal intensities could be directly compared. The statistically significant difference observed between gmIs28 and crml-1; gmIs28 is marked (†, P<0.009); n.s., not significant. Error bars indicate s.e.m. At least 80 animals were scored per genotype. The increase observed in the crml-1 mutant background was confirmed through an independent experiment that was scored blindly (data not shown). (H) Relative fluorescence intensities of SAX-3::GFP signals in ALM and BDU neurons from G. Only animals with both cells clearly in focus were included; at least 30 animals were scored per genotype. Error bars indicate s.e.m. Statistically significant differences between gmIs28 alone and the two unc-73-containing strains are marked (#, P<0.03).