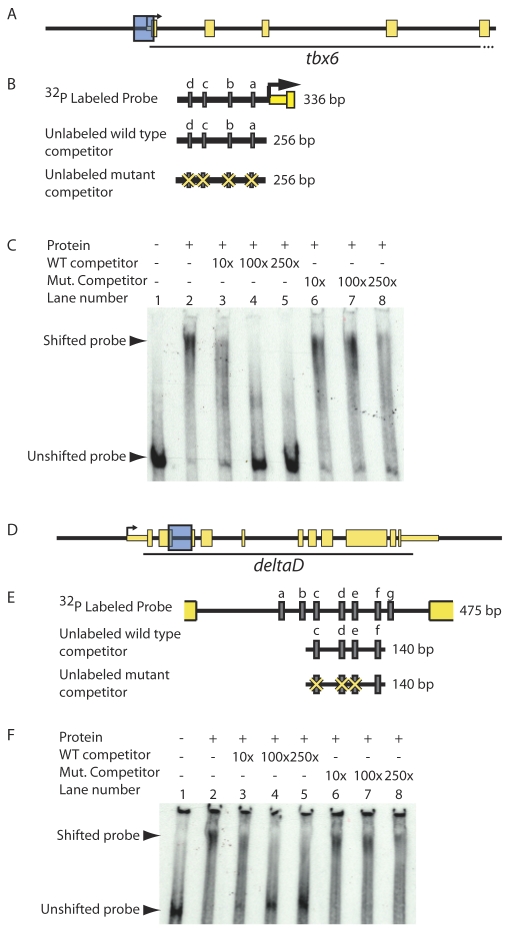
Fig. 5.
T-box proteins bind putative regulatory sequences in vitro in a T-box binding site-dependent manner. (A-F) To test whether Spt and Ntl bind putative regulatory sequences in vitro, we performed gel shifts using equal volumes of recombinant Spt-GST and Ntl-GST, and radiolabeled DNA corresponding to putative tbx6 and dld genomic regulatory sequences (shown as blue boxes in A and D and magnified in B and E to indicate Spt/Ntl binding motifs a-d and a-g, respectively, as grey rectangles). Spt and Ntl bind a 336 bp region containing four T-box sites just upstream of tbx6 (C; lane 1 versus lane 2). Addition of 10-250× excess unlabeled competitor competes well for binding and virtually eliminates DNA-protein complex formation (C; lanes 3-5); however, excess unlabeled competitor containing mutated T-box sites competes poorly (C; lanes 6-8). Similarly, a mixture of equal volumes of Spt-GST and Ntl-GST binds a 475 bp fragment from the dld second intron containing seven putative T-box sites (F, lane 1 versus 2). Unlabeled competitor containing four of the T-box sites competes for binding (F; lanes 3-5), but fails to compete when three of the T-box sites are mutated (F, lanes 6-8).