Fig. 2.
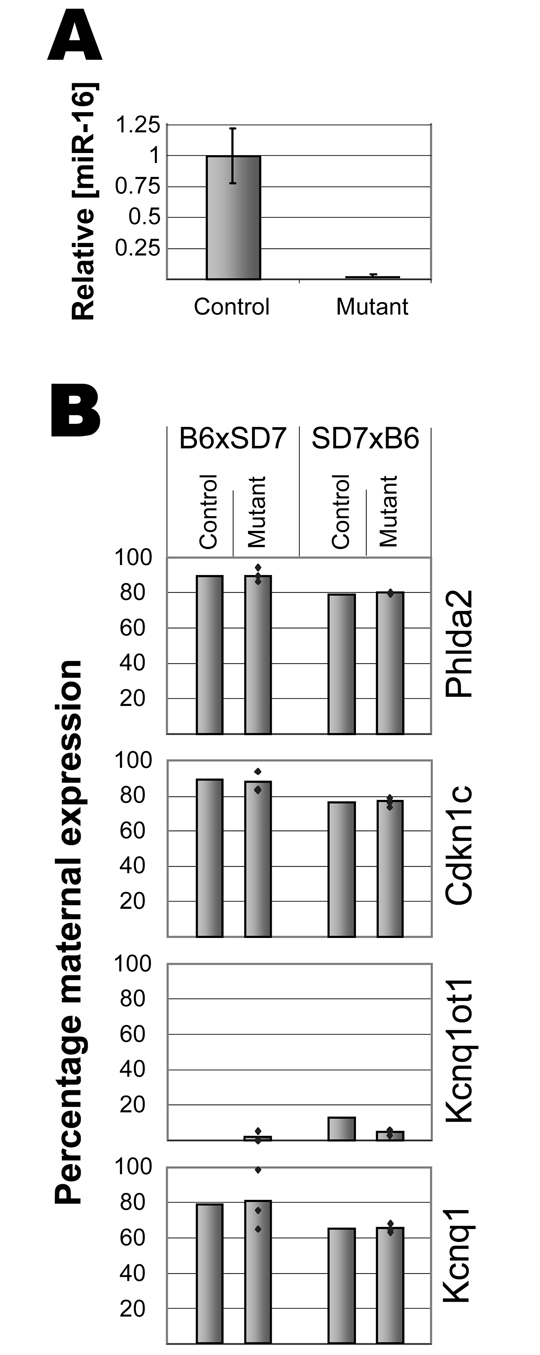
Silencing by Kcnq1ot1 is not dependent on the RNAi pathway. (A) qRT-PCR analysis of miR-16 abundance in control (Dcrflox/Δ; Meox+/+) E10.5 and mutant (Dcrflox/Δ; MeoxCre/+) E10.5 embryos, normalised to SnoRNA-202. The mature form of miR-16 is present in the mutants at a 50-fold lower level, demonstrating that Dicer action has been drastically reduced in the mutants. Error bars represent s.d. (B) RT-PCR and Sequenom MassArray analysis was performed to determine the percentage of maternal expression for the imprinted genes Phlda2, Cdkn1c, Kcnq1ot1 and Kcnq1 in E10.5 embryos. Means are represented as bars and individual data points as blue dots (one control embryo is shown for each cross). Data are shown for both the B6xSD7 and the reciprocal cross to exclude effects due to sequence variations between the B6 and SD7 alleles. Dicer mutants did not show loss of imprinting.
