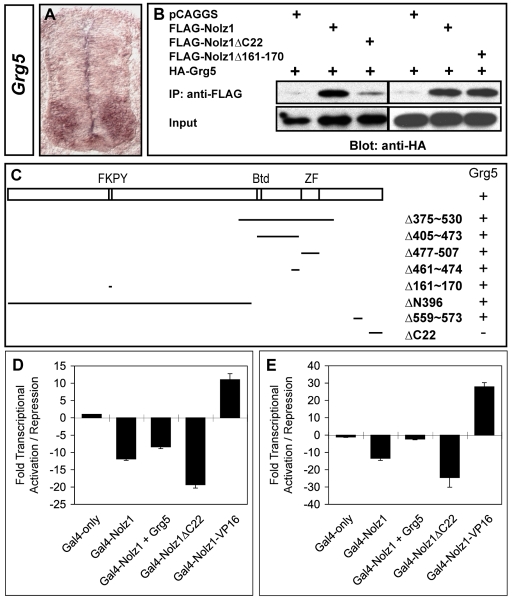
Fig. 4.
Nolz1 and Grg5 interact and function as transcriptional repressors. (A) In situ hybridization of Grg5 mRNA expression in transverse sections of chick embryonic spinal cords. (B) Co-immunoprecipitation (IP) analyses of Grg5 protein interactions with Nolz1. (C) Schematic of the Nolz1 coding region. FKPY, putative Grg consensus binding site; Btd, Buttonhead domain; ZF, atypical zinc-finger domain. Lines beneath indicate the regions deleted within the Nolz1 coding sequence; the numbers indicate the corresponding amino acids that are deleted. Whether Nolz1 retains (+) or loses (-) its ability to interact with Grg5 in co-IP is indicated alongside. (D) Luciferase-based transcriptional assays using extracts from transfected COS-7 cells. Data are from three replicate experiments, mean ± s.e.m. Student's t-test compared with Nolz1: Nolz1+Grg5, P=0.0055; Nolz1ΔC22, P=0.0016. (E) Luciferase-based transcriptional assays using extracts from electroporated chick spinal cords. n=6-8 embryos, mean ± s.e.m. Student's t-test compared with Nolz1: Nolz1+Grg5, P<0.000001; Nolz1ΔC22, P=0.0029. Compared with GAL4-only: Nolz1+Grg5, P=0.0003.