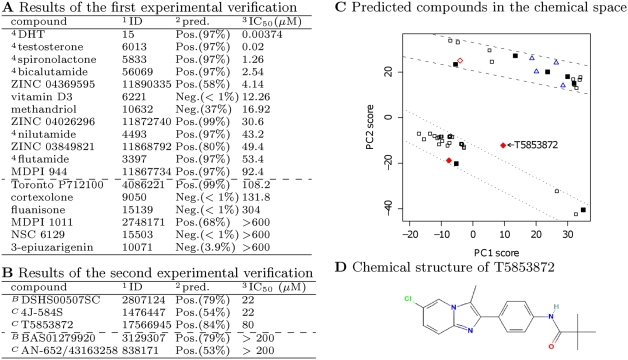
Figure 2. The first and the second experimental verifications showed more than 60% accuracy of computational predictions and the chemical space of verified compounds was explored.
(A) Results of the first in vitro binding assay. (B) Results of the second in vitro binding assay. (C) The chemical space based on E-Dragon [14] descriptors and the principal component analysis (PCA) applied to known ligands and additional data. The black squares correspond to known ligands in the training data, the solid black squares represent known approved drugs, the blue triangles correspond to true positives in the first computational prediction, and the red diamonds correspond to true positives in the second computational prediction. The open red diamonds belong to Figure 1B, and the solid red diamonds belong to Figure 1C. Chemical compounds located between the two dashed lines have steroid-like structures. (D) A potential ligand with a chemical structure differing from the structures of known ligands. In (A) and (B), 1; PubChem Compound ID. 2; computational prediction expressed as “label (predicted probability for a positive outcome)”. 3; The concentration of an unlabeled test compound, in which, according to the measured radioactivity, 50% of the [3H]-DHT is still bound to MBP-ARC. 4; chemical compounds included in the DrugBank set or additional data. B (C); predictions belonging to Figure 1B (C).