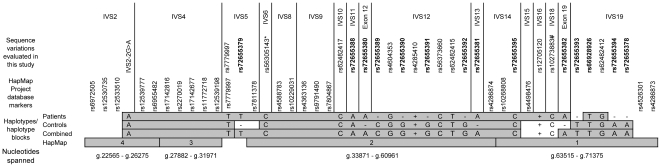
Figure 1. Haplotype analysis of patients and controls compared to International HapMap Project database markers.
Haplotype blocks as determined by HaploView 4.0 are shown as shaded bars and compared for patients, controls, patients and controls combined (Combined), and the International HapMap Project database markers (HapMap). The location of markers within the SLC26A5 gene is shown at the top of the figure. DNA sequence variations are shown above the haplotype blocks. Note: The only marker observed in this study that is included in the International HapMap Project database is rs7779997 in IVS5. The genomic SLC26A5 nucleotides spanned by the haplotype blocks derived from the International HapMap Project database markers are shown at the bottom of the figure. Abbreviations and symbols used: IVS, intervening sequence (intron); *formerly hCG1811409, Celera database [17]; #, C allele noted is variant allele, not reference sequence allele; −, Variant allele not detected; +, Reference allele not detected; 1, 2, 3, 4, represent haplotype block designations derived from and assigned by HaploView 4.0 software analysis of the International HapMap Project database markers. The reference sequence (undeleted) allele of variant rs66928926 (g.69917_69919delTCT) is designated as “T” and of variant rs72655394 (g.70078_70082delATATA) is designated as “A.” Novel variants are shown in bold type.