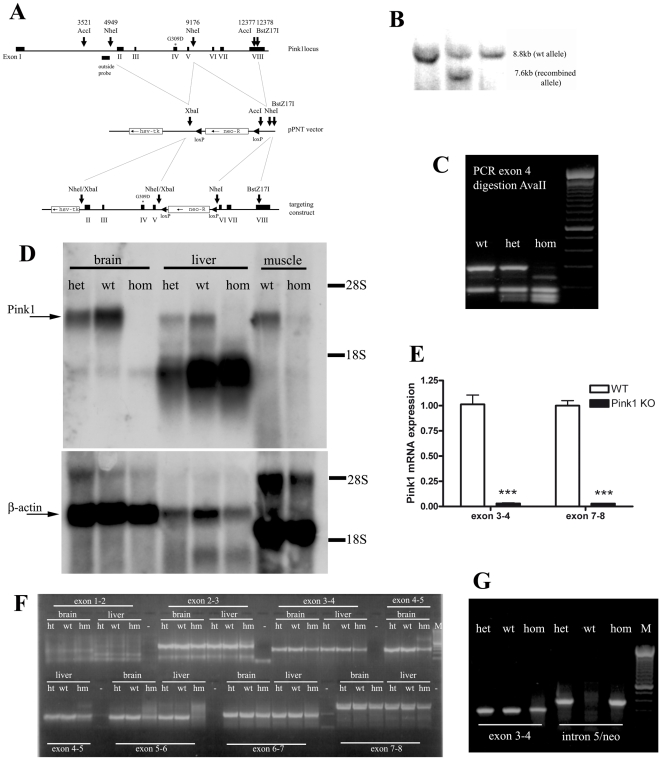
Figure 1. Generation and characterization of the Pink1 −/− mouse.
(A) Schematic drawing of the targeting strategy. (B) Demonstration of the allele representing the homologous recombination event at the Pink1 locus by Southern blotting of the outside probe in the embryonal stem cell line, (C) Presence of mutation in homozygous (hom) and heterozygous (het) mouse tail DNA represented by a double band (134+154 basepairs) instead of the wildtype (wt) single band (288 bp) after exon 4 amplification and AvaII restriction. (D) Absence of Pink1 transcript expression in three tissues of homozygous mutant versus heterozygous and wildtype mice by Northern blotting, using β-actin as control for equal loading and the 18S and 28S ribosomal bands as references for size. One additional cross-reacting band showed intensities not correlating with the mutant genotypes. No residual Pink1 transcripts of different sizes were detectable, indicating instability and degradation of the mutant mRNA. (E) 97% reduction of Pink1 mRNA in homozygous KO mice (n = 5) in independent TaqMan systems detecting sequences upstream and downstream from the Neo insertion site. (F) Saturation RT-PCR amplification of Pink1 mRNA sequences containing each splice site, studying brain and liver mRNA from wildtype, heterozygous (ht) and homozygous (hm) mice, demonstrating abnormalities in homozygous tissues with additional large bands for the exon 4–5 splicing boundary and smears of abnormally large size for the exon 5–6 boundary. (G) Saturation RT-PCR with primers from Pink1 intron 5 and the Neo selection marker demonstrating the pathological presence of intronic and Neo sequences in Pink1 mRNA from mutant (ht and hm) mouse samples.