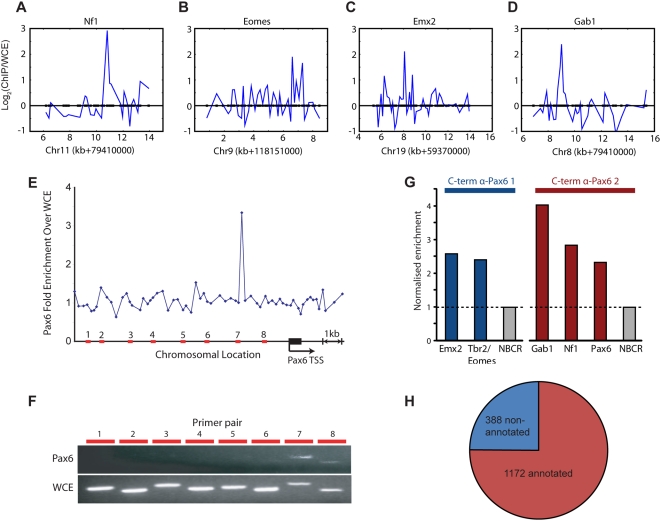
Figure 2. In vivo location analysis identifies the set of genes whose promoters are bound by Pax6 in neocortical stem and progenitor cells.
(A–D) Identification of Pax6-bound promoters. ChIP array data for four typical Pax6-bound promoters, all of which were calculated as significantly bound above background levels (see Materials and Methods for details). Each graph plots fold-enrichment (in log base 2) on the y-axis, against relative chromosomal position on the x-axis. (E,F) Validation of the specificity and accuracy of the binding of the Pax6 promoter by Pax6, confirmed by qPCR in (G). Scanning PCR primers were designed to amplify DNA from a series of genomic regions in the Pax6 promoter (red bars represent amplified regions in the plot of fold-enrichment, y-axis, against relative chromosomal position, x-axis). The peak of Pax6 binding from the ChIP array analysis is shown in E. Pax6 binding was confirmed in this assay only in the region around the oligonucleotide, as shown in the gel images in F. Pax6, Pax6 ChIP; WCE, whole cell extract. (G) Validation of Pax6-binding to a set of promoters by quantitative PCR in independent ChIPs (not used for the array hybridisations) using two different Pax6 polyclonal antibodies. Enrichment of a region is calculated relative to that observed in control ChIPs without the primary antibody and normalised to a non-bound control region (NBCR). (H) Three independent ChIPs and promoter array hybridisations were carried out for Pax6, in which Pax6-bound material was compared with total genomic DNA (whole cell extract, WCE) to identify regions enriched in the Pax6-ChIP material by raw ratio measurements. Pax6 is found on the promoters of 1560 genes, 1172 (75%) of which are annotated genes (Table S1).