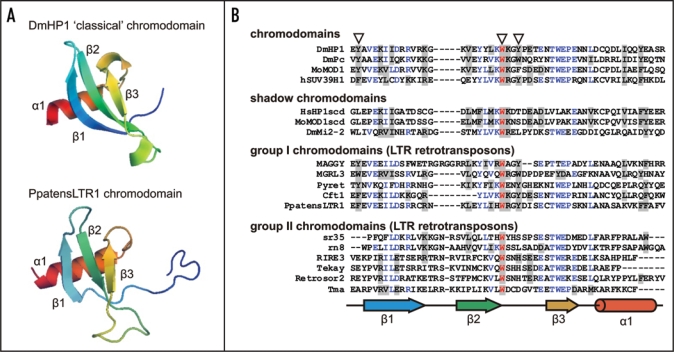
Figure 1.
Structures of chromodomains from the HP1 protein (Drosophila melanogaster; pdb:1q3l)4 and PpatensLTR1 (Gypsy LTR retrotransposon from moss Physcomitrella patens) (A); and multiple alignment of ‘classical’ chromodomains and ‘shadow’ chromo-like motifs from functional proteins as well as group I and group II Gypsy LTR retrotransposon chromodomains (B). The PpatensLTR1 chromodomain structure was predicted by (PS)2.5 The residues that constitute the aromatic pocket are indicated by arrows at the top. The secondary structural elements are shown at the bottom: arrows indicate β-strands; cylinder indicates α-helix. The GenBank accession numbers for the sequences are as follows: DmHP1—M57574; DmPc—P26017; MoMOD1 and MoMOD1scdv—P23197; hSUV39H1—S47004; HsHP1scd—AAB26994; DmMi2-2—AF119716; MAGGY—L35053; MGRL3—AF314096; Pyret—AB062507; Cft1—AF051915; PpatensLTR1—XM_001752430; sr35—AC068924; rn8—AK068625; RIRE3—AC119148; Tekay—AF050455; RetroSor2—AF061282; Tma—AF147263.