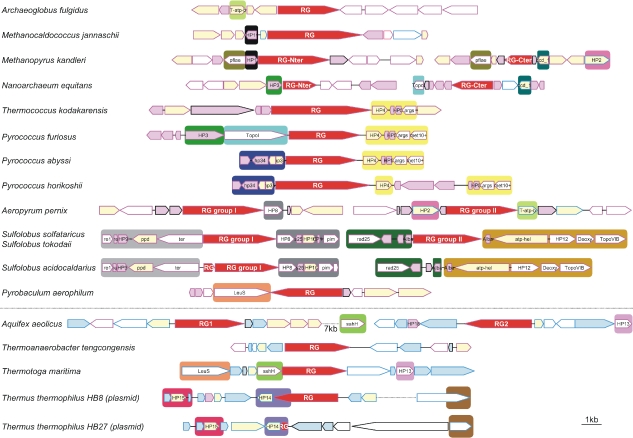
Figure 1.
Genomic organization of the regions surrounding reverse gyrase genes. Blue boxes indicate genes having exclusively bacterial or bacterial and eukaryotic homologues; pink boxes indicate genes having exclusively archaeal or archaeal and eukaryotic homologues; yellow boxes indicate genes having both archaeal and bacterial homologues; and empty boxes indicate genes with homologues in the three domains of life. The color of the box frames indicates the nature of each gene's closest homologue by BLASTP searches (blue = bacterial; and pink = archaeal). Large boxes with the same color surrounding genomic fragments indicate regions sharing homologous genes. The scale bar represents approximately 1000 nucleotides. Gene name abbreviations: T-atp-b = ABC transporter, ATP-binding protein (COG1131); HP1 = hypothetical protein (COG1590); pflae = pyruvate-formate lyase-activating enzyme (COG1180); dcd_1 = deoxycytidine deaminase (COG0717); HP2 = uncharacterized membrane protein (COG3174); HP3 = hypothetical protein (COG2433); TopoI = DNA topoisomerase I (COG0550); HP4 = predicted transcriptional regulators (COG1378); HP5 = uncharacterized Zn-finger containing protein (COG1645); args = arginase 1-like protein (COG0010); met10+ = met-10+ protein (COG2520); HP6 = uncharacterized membrane-associated protein/domain (COG2512); HP7 = predicted DNA-binding proteins with PD1-like DNA-binding motif (COG1661); HP8 = predicted amidohydrolase (COG0388); re1 = DNA-directed RNA polymerase, subunit E (COG1095); re = DNA-directed RNA polymerase, subunit E (COG2093); HP9 = hypothetical protein (COG1909); ppd = phosphonopyruvate decarboxylase (COG3635); ter = transitional endoplasmic reticulum ATPase (COG0464); s25 = SSU ribosomal protein S25E (COG4901); HP10 = membrane protein implicated in regulation of membrane protease activity (COG1585); HP11 = predicted transcriptional regulators containing the CopG/Arc/MetJ DNA-binding domain (COG3609); pim = protein L-isoaspartate methyltransferase (COG2519); rad25 = DNA repair protein rad56 (COG1061); Alba = archaeal DNA-binding protein (COG1581); atp-heli = ATP-dependent helicase (COG1201); HP12 = hypothetical protein (COG1690); Deoxy = deoxyhypusine synthase (COG1899); TopoVIB = DNA topoisomerase VI subunit B (COG1389); LeuS = leucyl-tRNA synthetase (COG0495); sahH = adenosylhomocysteinase (COG0499); HP13 = hypothetical protein (COG1578); HP14 = predicted metal-dependent membrane protease (COG1266); HP15 = hypothetical protein (COG1604); and HP16 = hypothetical protein (COG1458).