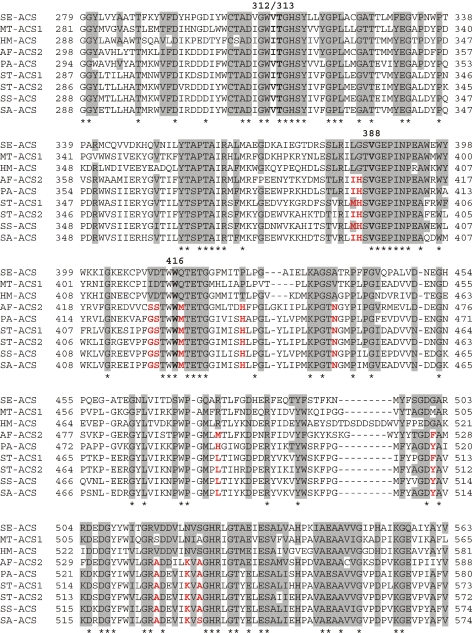
Figure 4.
Alignment and ConSurf analysis of ACS sequences. The M. thermautotrophicus MT-ACS1 and A. fulgidus AF-ACS2 amino acid sequences were aligned with the S. enterica ACS sequence (SE-ACS) and the ACS sequences from P. aerophilum (PA-ACS), Sulfolobus tokodaii (ST-ACS1 and ACS2), Sulfolobus solfataricus (SS-ACS), and Sulfolobus acidocaldarius (SA-ACS) using Clustal X (Thompson et al. 1997). A partial alignment is shown here. Residues of the S. enterica ACS found to have high evolutionary conservation scores by ConSurf analysis (Armon et al. 2001, Glaser et al. 2003, Landau et al. 2005) (http://consurf.tau.ac.il) are shaded, as are residues in the other ACS sequences that are identical or among the alternative residues listed for each of these highly conserved positions. Asterisks indicate those positions that are identical in all nine sequences. The acetate binding pocket residues are boldfaced and numbered above the aligned sequences according to their position within MT-ACS1. Those residues at positions with high ConSurf scores that differ in AF-ACS2, PA-ACS, and the Sulfolobus sequences from the three ACS sequences representing enzymes with “traditional” characteristics are indicated in red.