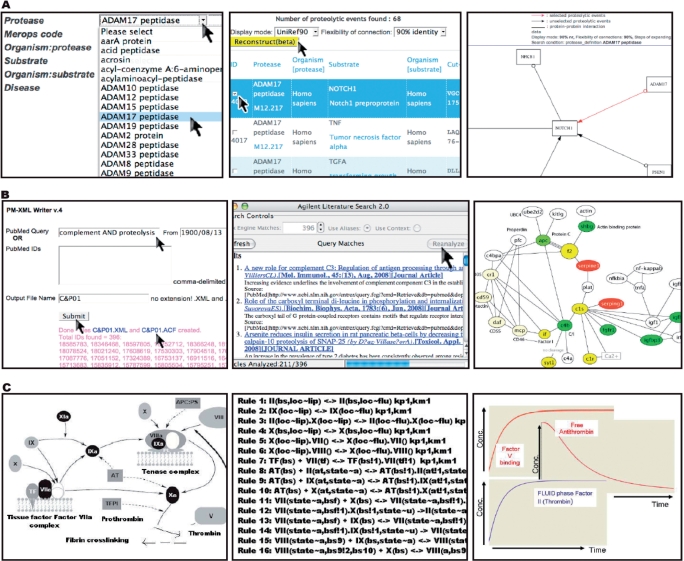
Figure 3.
Three types of information in PathwayDB. (A) Automated network reconstruction for one proteolytic events caused by ADAM17 peptidase and Notch1 preproprotein. Left panel: from CutDB front page, users search for the events. Middle panel: selection of events to reconstruct networks. Users can also define three parameters to show the network diagrams: (i) ID display mode; (ii) flexibility of connection, the nodes are connected within given threshold of sequence similarity; and (iii) steps of expanding, the edges are extended by given extension level. Right panel: the reconstructed pathway. The red arrows indicate the selected events. The black arrows indicate expanded events. The nonarrows edges are protein–protein interactions. (B) Proteolytic inferred networks. Left panel: PubMed-XML writer, a web service for extracting Medline abstracts, is used to delimit lexical queries, for example ‘complement AND proteolysis’. These abstracts are then assembled in XML format and submitted to Agilent Literature Search, acting as plug-in to Cytoscape (middle panel), which parses sentences containing ontologically known entities (such as gene and protein names). Cytoscape takes these gene/protein names and builds interactive networks as hyper-graphs. Right panel: hyper-graph generated for one ‘Network of the month’ entitled ‘Complement activation and regulation 01’. This network captured five proteases (nodes in yellow) that interplay with six substrates (nodes in green), four cofactors (nodes in light yellow), two inhibitors (nodes in red) and other binding proteins such as C4BP (white nodes). (C) Rule-based modeling of network function. Left panel: schematic representation of major events involved in coagulation. Middle panel: rules for coagulation cascade. Right panel: modeling of production/consumption profiles of individual components of the pathway using an in-house developed rule-based model for coagulation. In the plot, the x-axis denoted the time of simulations (100 iterations) and the y-axis is the molecular concentrations in nanomolar (0–100 000).