Abstract
The UCSC Genome Browser Database (GBD, http://genome.ucsc.edu) is a publicly available collection of genome assembly sequence data and integrated annotations for a large number of organisms, including extensive comparative-genomic resources. In the past year, 13 new genome assemblies have been added, including two important primate species, orangutan and marmoset, bringing the total to 46 assemblies for 24 different vertebrates and 39 assemblies for 22 different invertebrate animals. The GBD datasets may be viewed graphically with the UCSC Genome Browser, which uses a coordinate-based display system allowing users to juxtapose a wide variety of data. These data include all mRNAs from GenBank mapped to all organisms, RefSeq alignments, gene predictions, regulatory elements, gene expression data, repeats, SNPs and other variation data, as well as pairwise and multiple-genome alignments. A variety of other bioinformatics tools are also provided, including BLAT, the Table Browser, the Gene Sorter, the Proteome Browser, VisiGene and Genome Graphs.
INTRODUCTION
The UCSC Genome Browser Database (GBD, http://genome.ucsc.edu) provides access to the DNA sequences for the human genome and many other organisms (1–4). The database also contains annotation datasets for a wide variety of data types aligned to the reference genome sequence, which are displayed graphically as ‘tracks’ in the UCSC Genome Browser. Currently, the GBD offers sequence, annotations and browsers for 14 mammals, 10 nonmammalian vertebrates and 22 invertebrates, including 11 Drosophila species and six worms. Although we do not provide browsers for low-coverage assemblies, the GBD incorporates the sequences of bushbaby, treeshrew, rabbit, common shrew, hedgehog, armadillo, elephant and tenrec into the human and mouse comparative genomic annotations. We add new and updated assemblies to the database as they are released by the sequencing centers, and maintain older assemblies either on the main site or in the genome archives (http://genome-archive.cse.ucsc.edu), where the complete history of the human genome sequence is available. Links to other major genome databases, including Ensembl (5) and NCBI MapViewer (6), are provided throughout the site.
Genome assemblies are annotated with assembly clone details, GenBank mRNAs (7), RefSeq alignments (8), microarray gene expression data, regulatory element tracks, SNP and other variation data, multiple genome alignments and other datasets. The annotations offered in the Genome Browser's Comparative Genomics track group facilitate navigation among organisms using both the pairwise alignments in the chain and net tracks and multiple alignments (multiz) (9).
Data in the GBD are updated regularly, including nightly updates of new mRNA submissions to GenBank (alignments of all new sequences to all assemblies), MGC (10) and consensus coding sequence (CCDS); weekly updates of EST data; and a complete realignment whenever GenBank releases a periodic update. Certain other datasets are also updated regularly via new automated processes, including Ensembl genes annotations (5) on several organisms (updated 3–5 times a year), monthly updates of mouse data from the International Gene Trap Consortium (IGTC) (11) and regular new releases of the Database of Genomic Variants (DGV) (12). By providing up-to-date releases of data originated by other groups, along with convenient linkouts to the primary sources, we seek to maintain our database as an integrated resource for the scientific community. All data are freely available via the Genome Browser and Table Browser interfaces, and may be downloaded in bulk at http://hgdownload.cse.ucsc.edu. The source code and binaries are free for noncommercial use.
In addition to the Genome Browser graphical interface, the GBD provides other tools for efficient data mining. The Table Browser (13) continues to be one of the most widely used features of the GBD toolset and is increasingly used to export data to the Galaxy (14) tools at Penn State for further processing. The Gene Sorter (15), the Proteome Browser (16), VisiGene (3), Genome Graphs (4) and BLAT (17) have been previously described.
UCSC is the Data Coordination Center for the Encyclopedia of DNA Elements (ENCODE) project (18), which uses the GBD and Genome Browser for data storage and graphical access to the data. This project uses a variety of techniques to generate genome-wide annotations, including DNase hypersensitivity sites, mRNA expression, histone modification, transcription factor binding sites and gene annotations (Gencode). Data deposited for the ENCODE pilot project (now completed) are presented in the Genome Browser as separate track groups on the human hg18 assembly. Initial ENCODE production-phase data will become available in the coming year.
The evolving set of tools associated with the GBD has ever-increasing capability and configurability. Users can find assistance in using the database and tools via a large number of online help pages (http://genome.ucsc.edu/goldenPath/help), FAQs (http://genome.ucsc.edu/FAQ) and links to tutorials produced by Open Helix (http://openhelix.com). We also provide staff resources to address questions from users through our mailing list (genome@soe.ucsc.edu).
NEW DATA
New assemblies
Since the last GBD update paper was written in September 2007 (4), we have added 13 new genome assemblies to the database, including the initial assemblies for nine new organisms (orangutan, marmoset, guinea pig, zebra finch, lamprey, lancelet, and three Caenorhabditis species: brenneri, remanei and japonica) and updated assemblies for the cow, zebrafish, sea urchin and C. elegans. We provide a basic set of annotations for each new assembly, as well as alignments of GenBank data and pairwise alignments (chain and net tracks) (19) of the assembly to selected other organisms. Seven of the new assemblies have multiple-alignment annotations, including a comparison of six worm species on the latest C. elegans (ce6) browser.
New annotations
In addition to new assembly releases, more than 200 annotations have been added to existing genome assemblies in the past year. These annotations represent a wide variety of data types, including new microarray data for several organisms and a collection of variation data in the human assemblies (see below). This section summarizes a representative sample of the new annotation data. Further details on the construction of any annotation are easily obtained by clicking on an item in the corresponding track in the Genome Browser.
A new annotation on the hg18 human assembly, Pos Sel Genes in the Genes and Gene Prediction track group, shows genes under positive selection in one or more of six mammals (20). The track displays the results of a genome-wide scan for positively selected genes based on multiple alignments of the human, chimp, macaque, mouse, rat and dog genome assemblies. Orthologous genes were examined for evidence of positive selection using a series of nine likelihood ratio tests (LRTs) based on Yang and Nielsen's (21) branch-site framework.
New data from the Open Regulatory Annotation (ORegAnno) project show gene regulation annotations for four model organisms (human, mouse, Drosophila melanogaster, and yeast) (22). An ORegAnno record describes an experimentally proven and published regulatory region (promoter, enhancer, etc.), transcription factor binding site, or regulatory polymorphism. Each ORegAnno annotation has links to the ORegAnno database.
The human assembly now contains annotation data (the HGSV Discordant track) from Kidd et al. (23) that maps clones from eight individuals from the HapMap Project (24) to the reference assembly. This annotation shows regions where the known size of the clone does not match the size of the reference, representing a putative large indel, and provides a valuable source of information and cloned DNA for mapping human genetic variation.
A 30-vertebrate alignment Conservation track is now available on the mm9 mouse assembly. This track, which displays the results of a multiz alignment and phastCons computation (25), is useful for viewing the evolutionary relatedness of sequences across a wide range of animals. We have also added a dataset to the mm9 assembly showing microRNAs from miRBase at the Wellcome Trust Sanger Institute (26).
On the rat rn4 assembly we now provide quantitative trait locus (QTL) data from the RGD (27). These data define more than 1000 loci in the rat genome that affect a phenotypic trait in a continuously distributed fashion, such as blood pressure and glucose level.
A new track of more than 7500 gene insertions in D. melanogaster (GDP Insertions) is displayed on the dm3 genome assembly. These annotations allow identification of genes for which P-element and Minos insertion strains are available from the Gene Disruption Project (28), with a direct link to the stock center in Bloomington for detailed information and ordering.
New UCSC Genes
In September 2008, we released a new version of the UCSC Genes dataset for the hg18 human assembly. The UCSC Genes annotation includes multiple isoforms of known protein-coding and noncoding genes based on a variety of criteria, including evidence from RefSeq, UniProt (29), GenBank and comparative genomics.
The latest UCSC Genes annotation uses the CCDS protein to determine the proper alignment where the CCDS and RefSeq are not in perfect agreement. At the time when we made this decision, it was our belief that the benefits of an international consensus outweighed minor differences in gene models resulting from arbitrary choices of alignment in tandemly duplicated regions and minor differences in opinion on the true 5′-end of a transcript. This has led us to choose CCDS over RefSeq for start codons or splice sites of 353 genes, for example, the splice junctions between exons 4 and 5 in the gene IFI35 (at hg18 chr17:38,418,889-38,419,044, http://genome.ucsc.edu/cgi-bin/hgTracks? db=hg18&position=chr17:38418889-38419044&knownGene=pack&refGene=pack).
The new UCSC gene set contains 66 803 genes (including isoforms) of which 13 767 are nonprotein-coding genes (Table 1). The genes are found in 26 570 clusters.
Table 1.
Summary of new UCSC Genes track
| UCSC Genes | Genes | Clusters | Previous | Change |
|---|---|---|---|---|
| Coding | 53 036 | 20 409 | 20 433 | −24 |
| Noncoding | 13 767 | 6161 | 5871 | +290 |
| Total | 66 803 | 26 570 | 26 304 | +266 |
This update includes links in the Genome Browser to and from external databases for the orthologous genes in several model organisms: the Mouse Genome Database, MGD (30), the Rat Genome Database, RGD (27), Zebrafish Information Network, ZFIN (31), WormBase (C. elegans) (32), FlyBase (Drosophila) (33) and Saccharomyces Genome Database (34). We plan to continue providing regular updates of the UCSC Genes track for the latest human and mouse assemblies.
To view the UCSC Genes annotation for a specific gene using the Genome Browser, type a search term into the Position box on the Genome Browser webpage. It is possible to search on a wide variety of gene identifiers, such as HGNC names (35) or UniProt ID as well as keywords from the GenBank or UniProt descriptive text. The latter approach will also find genes whose products are associated with each other, provided the association is annotated in the RefSeq text.
The details pages for the UCSC Genes track contain links to local resources such as the Gene Sorter, Proteome Browser and the VisiGene in situ hybridization image archive as well as links to a wide variety of external databases. New linkouts this year include Human Cortex Gene Expression data from the Allen Brain Institute, Human Genome Epidemiology (HuGE) data (36) and the Comparative Toxicogenomics Database (CTD) (37).
Variation
The hg18 human assembly offers a number of human variation annotations, several of which have been updated in the past year. Of particular note, we have added SNP data from dbSNP release 129 (38) to supplement the existing dbSNP data from the 128 and 126 releases on the human hg18 assembly.
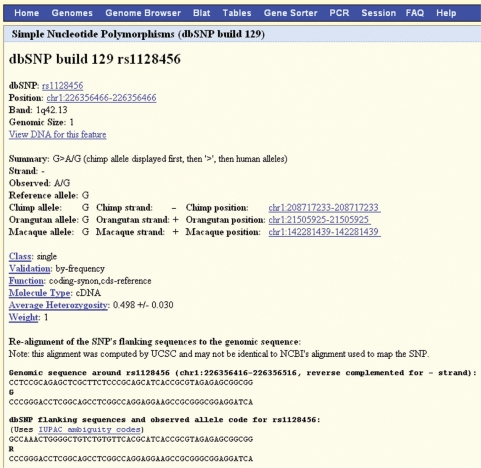
The Genome Browser details pages for the SNP 129 annotation track have been expanded to capture much of the data from the dbSNP database, including the type of SNP (coding, noncoding, synonymous, etc.). We now also display the alignment to the reference assembly of the region surrounding the SNP. Additionally, for comparative purposes, the orthologous alleles from several primate species (chimp, orangutan and rhesus) are given. Figure 1 shows part of the SNP 129 details page for SNP rs1128456.
Figure 1.
SNP 129 track details page showing partial information about SNP rs1128456 on chromosome 1.
We have also updated the mm9 SNP annotation to dbSNP version 128 and have released dbSNP 127 on the bosTau3 cow assembly.
A new annotation track in the Comparative Genomes group on the hg18 assembly, Cons Indels MmCf, uses evolutionary conservation among human, mouse and dog to identify small indels in the human reference assembly. Other new variation data tracks on hg18 (all in the Variation and Repeats track group) include DGV Structural variants, Segmental Dups, Exapted Repeats and Interrupted Repeats.
UCSC has removed data from the Wellcome Trust Case Control Consortium study and the NIMH study of bipolar disorder in response to the policy decision by NIH that these data could potentially be used to identify individuals under certain circumstances, in possible violation of the terms of consent for the studies. We will continue working with other groups in the international research community to determine how best to protect the confidentiality of participants of genome-wide association studies (GWAS), while making these data accessible for scientific research. Depending on the outcome of these discussions, we plan to provide more GWAS data in the Genome Browser in the future, as well as new graphical tools for viewing and analyzing clinical trial data.
Transmap
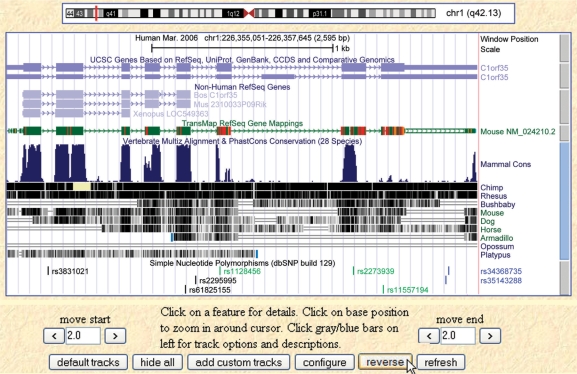
A group of new data tracks, known collectively as TransMap, has been added to all vertebrate genome assemblies in the Genes and Gene Prediction group. These tracks map genes and related annotations in one species to another, using synteny-filtered pairwise genome alignments (chains and nets) to determine the most likely orthologs. Individual tracks within the TransMap supertrack include mappings based on, respectively, mRNA, RefSeq or UCSC Genes evidence (39). For the mRNA TransMap track on the human assembly, for example, more than 400 000 mRNAs from 23 vertebrate species were aligned at high stringency to the native assembly using BLAT. The alignments were then mapped to the human assembly using the chain and net alignments produced using Blastz (40), which has higher sensitivity than BLAT for diverged organisms. Compared with translated BLAT (Non-Human RefSeq Genes, Figure 2), TransMap finds fewer paralogs and aligns more UTR bases (Figure 2). For closely related low-coverage assemblies, a reciprocal–best relationship is used in the chains and nets to improve the synteny prediction. As with all GBD annotations, the details of the dataset construction may be found on the corresponding Genome Browser track details pages.
Figure 2.
Screen image of Genome Browser for hg18 human assembly, chromosome 1, showing several new features. From top to bottom: Scale bar; UCSC Genes track; Non-human RefSeq Genes; TransMap RefSeq Genes, showing improved mapping of bases at 3′-end of a mouse RefSeq (red) that did not map in Non-Human RefSeq track; Conservation track; SNP 129 track. Entire image has been reversed from default configuration using Reverse button (cursor arrow at bottom) to show Transmap annotations in 5′-to-3′ direction.
New Gene Sorter columns
The Gene Sorter allows users to sort genes using a variety of criteria, including expression pattern or protein homology, with a large number of user-specified data fields displayed in columns for each gene. The tool provides convenient links back to the Genome Browser or to the gene description on the UCSC Gene details pages, as well as expression profiles, protein–protein interaction data and others. Several new columns have been added to the Gene Sorter in the past year for the six model organisms supported by the Gene Sorter: human, mouse, rat, C. elegans, D. melanogaster and yeast.
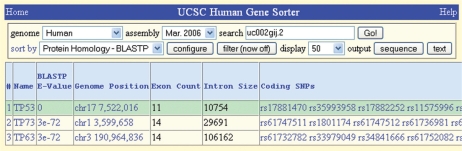
The Intron Size column displays the largest or smallest intron for each gene; the Coding SNPs column gives convenient access to exon polymorphism information; CDS Score shows a computation of the likelihood that the gene encodes a protein; Gene Category classifies the gene as coding, noncoding, antisense, etc. and Exon Count records the number of exons (Figure 3).
Figure 3.
Gene Sorter output showing new columns for the TP53 gene (top row) and all genes meeting the criterion, Protein Homology–BLASTP. Columns shown, left to right: BLASTP E-value, Genome Position, Exon Count, Intron Size (set to maximum size) and Coding SNPs (truncated).
NEW DISPLAY FEATURES
We have added a number of new display features to the Genome Browser in the past year, many of which are usability improvements based on feedback from the research community. The Base Position track now provides an optional automatic scale bar configurable through its description page. A Reverse button below the main browser image allows users viewing genes that align on the negative strand to flip the entire display so that the gene of interest appears in the 5′-to-3′ direction (Figure 2). It is now possible to navigate directly to a single nucleotide by typing the coordinate into the Position box; e.g., chr1:226356466 will locate the SNP rs1128456. (Note that SNPs can still be accessed directly by typing the rs number into the box.)
Several enhancements have been added to the track groups below the main browser image. Track groups are now collapsible, allowing the user to hide groups that are not of interest. Tracks can be moved from one group to another, including to the Custom Tracks group at the top, allowing users to collect tracks of interest in one place for a more customized viewing experience. Each of the track group header bars now has a Refresh button that eliminates the need to scroll up or down the page to submit a change. A number of enhancements have been made ‘under the hood’ to improve the performance of the Genome Browser. To reduce the number of track controls beneath the browser image and to speed the refresh of the page, certain groups of related tracks have been combined into super-tracks that share configuration options. For example, the individual tracks within the TransMap track on vertebrate assemblies are controlled together. Track controls for super-tracks are distinguished by an ellipsis (…) in the label name.
The details pages of multiple alignment tracks now allow users to obtain DNA sequences from low-coverage assemblies for which no genome browser is provided. The in silico PCR function now creates a track on the Genome Browser image, allowing the user to visualize the relationship between an amplified fragment and other annotations, most usefully exons and introns. When the primers used to generate the amplicon do not match the reference assembly, the browser highlights the differences with red coloration.
Custom tracks enhancements
The custom track feature of the UCSC Genome Browser allows users to view their own data in the context of all the resident data on the browser. We have enhanced this feature in several ways. For example, the ‘next item’ feature, previously available only for selected UCSC-hosted tracks, is now available for a broader set of tracks including user-created custom tracks. This feature allows the user to quickly move to the left or right in the browser display to the next feature in a particular track, which is particularly useful for custom tracks with sparse coverage.
We have also extended the custom track feature to two new data types. The bedGraph data type provides a simplified method for displaying quantitative data in the browser. The MAF data type allows users to upload multiple-alignment data to the browser as a custom track. This will prove to be especially useful as high-throughput sequencing methods become more widely adopted.
The internal representation of custom tracks in the browser is now based on database tables on a dedicated machine rather than the previous file-based implementation. This offers a considerable speedup of the display, particularly when users revisit the browser in subsequent sessions.
One of the most popular tools introduced to the Genome Browser in recent years is the session-saving function, which allows users to save and share multiple browser configurations for future use (4). Custom tracks that are associated with saved sessions in the browser now have increased longevity. Typically, custom tracks are kept on the Genome Browser for 48–72 h after the last access. However, when users associate the tracks with a saved session (the ‘Session’ link in the top navigation bar), an effort is made to maintain the tracks for a minimum of several weeks. A related change now informs the user in the Session interface that there are custom tracks being saved, indicating the associated genome assembly.
Because there are many browser settings and an unlimited number of combinations of tracks and display options, the Genome Browser uses browser cookies that persist from one visit to another to maintain the state of user sessions. This allows users to revisit the browser on subsequent days and resume a session without having to reestablish their session configuration. Users frequently need to have more than one instance of the Genome Browser interface on their computer desktop simultaneously. The browser now has a formal method of preventing these instances from interfering with one another. When a user spawns a new browser instance, the new session inherits all the settings of the original, but thereafter maintains separate parameters, allowing independent browsing in several windows at once.
Future directions
UCSC will continue to add new vertebrate and selected invertebrate model organism assemblies and browsers to the GBD as the sequences become available. We are working closely with NCBI and Ensembl to standardize the process for obtaining and distributing new sequence data to ensure that all centers are offering the same versions. We expect to release a 44-species multiple alignment track for the 2×-coverage species project and an expanded multiple alignment Conservation track for the latest human assembly. Data from the 1000 Genomes project will be incorporated into the variation annotations, and will include high-resolution maps of recombination hotspots.
Several browser enhancements are planned, such as expanding the usability and configurability of our data-browsing tools, upgrading isPCR to allow users to query RNA space to align sequence separated by introns and adding support for custom tracks containing more than one type of data (mixed composite tracks). Within the next year we plan to release a new type of track that displays user input directly on the Genome Browser via a wiki mechanism, which will allow experts on particular genes to post comments, data references and other information directly on the UCSC site. Finally, UCSC has been developing the capacity to display access-controlled medical data, e.g., HIV genomics and clinical data, in collaboration with Global Solutions for Infectious Diseases. A new cancer genomics browser has been completed in collaboration with several research groups. We anticipate that a public version of it will be deployed after access and confidentiality issues are resolved.
FUNDING
The National Human Genome Research Institute (1P41HG002371-08 to UCSC Center for Genomic Science, 2P41HG002371-08 ENCODE supplement to UCSC Center for Genomic Science, 1P41HG004568-01 UCSC ENCODE Data Coordination Center, a subcontract on 2U41 HG004269-02 to L.Stein for A Data Coordination Center for modENCODE and a subcontract on 1U54HG004555-01 to T.Hubbard for Integrated Human Genome Annotation; Generation of a Reference Gene Set); National Cancer Institute (Contract No. N01-CO-12400 for Mammalian Gene Collection); the Howard Hughes Medical Institute (to D.H.). T.W. is a Helen Hay Whitney fellow. Funding for open access charge: the Howard Hughes Medical Institute.
Conflict of interest statement. R.M. Kuhn, D. Karolchik, A.S. Zweig, K. E. Smith, K. R. Rosenbloom, B. Rhead, B. J. Raney, A. Pohl, F. Hsu, A. S. Hinrichs, R. A. Harte, M. Diekhans, H. Clawson, G. P. Barber, D. Haussler and W.J. Kent receive royalties from the sale of UCSC Genome Browser source-code licenses to commercial entities.
ACKNOWLEDGEMENTS
We would like to thank the many collaborators who have contributed data to our project, our Scientific Advisory Board for their valuable advice and recommendations, and our users for their feedback and support. We would also like to acknowledge the dedicated system administrators who have provided an excellent computing environment: Jorge Garcia, Erich Weiler, Chester Manuel and Victoria Lin.
REFERENCES
- 1.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinrichs A, Karolchik D, Baertsch R, Barber G, Bejerano G, Clawson H, Diekhans M, Furey T, Harte R, Hsu F, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC Genome Browser Database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. The international gene trap consortium website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 13.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Miller W, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent WJ, Hsu F, Karolchik D, Kuhn RM, Clawson H, Trumbower H, Haussler D. Exploring relationships and mining data with the UCSC Gene Sorter. Genome Res. 2005;15:737–741. doi: 10.1101/gr.3694705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu F, Pringle TH, Kuhn RM, Karolchik D, Diekhans M, Haussler D, Kent WJ. The UCSC proteome browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Encode Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 19.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosiol C, Vinar T, da Fonseca R, Hubisz M, Bustamante C, Nielsen R, Siepel A. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 22.Griffith OL, Montgomery SB, Bernier B, Chu B, Kasaian K, Aerts S, Mahony S, Sleumer MC, Bilenky M, Haeussler M, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidd J, Cooper G, Donahue W, Hayden H, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The International HapMap Consortium. The international hapmap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, Dongen SV, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ, RGD_Team The Rat Genome Database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA, The Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. Wormbase 2007. Nucleic Acids Res. 2008;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson RJ, Goodman JL, Strelets VB, The FlyBase Consortium FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 37.Mattes WB, Pettit SD, Sansone S.-A, Bushel PR, Waters MD. Database development in toxicogenomics: issues and efforts. Environ. Health Perspect. 2004;112:495–505. doi: 10.1289/ehp.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherry S, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Sanborn J, Diekhans M, Lowe C, Pringle T, Haussler D. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput. Biol. 2007;3:e247. doi: 10.1371/journal.pcbi.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]