Abstract
We present a new database and an on-line search engine, which store and query the protein binding pockets that interact with single-stranded RNA nucleotide bases. The database consists of a classification of binding sites derived from protein–RNA complexes. Each binding site is assigned to a cluster of similar binding sites in other protein–RNA complexes. Cluster members share similar spatial arrangements of physico–chemical properties, thus can reveal novel similarity between proteins and RNAs with different sequences and folds. The clusters provide 3D consensus binding patterns important for protein–nucleotide recognition. The database search engine allows two types of useful queries: first, given a PDB code of a protein–RNA complex, RsiteDB can detail and classify the properties of the protein binding pockets accommodating extruded RNA nucleotides not involved in local RNA base pairing. Second, given an unbound protein structure, RsiteDB can perform an on-line structural search against the constructed database of 3D consensus binding patterns. Regions similar to known patterns are predicted to serve as binding sites. Alignment of the query to these patterns with their corresponding RNA nucleotides allows making unique predictions of the protein–RNA interactions at the atomic level of detail. This database is accessable at http://bioinfo3d.cs.tau.ac.il/RsiteDB.
INTRODUCTION
Understanding and predicting protein–RNA interactions at the atomic level is crucial for our ability to interfere with such processes as gene expression and regulation. Several works have classified protein–RNA interactions based on the sequences and folds of the corresponding protein (1–3) or RNA molecules (4–6). However, these do not always capture the similarity in the local regions which are responsible for protein–RNA recognition. These regions are important since even proteins of the same family can form different interactions with RNA nucleotides (7,8). By analyzing the amino acid composition in RNA binding sites, several successful methods for prediction of RNA binding regions were developed (9–12). However, there are only few methods (5) that distinguish between two main interaction types formed across protein–RNA interfaces: (i) interactions with the backbone of double-stranded RNA molecules; (ii) interactions with single-stranded RNA bases that are buried in the protein binding pockets (14).
Here, we focus on the classification and prediction of protein interactions formed with single-stranded nucleotide bases. Sequences of such nucleotides, which are not involved in local base pairs and are extruded from the surrounding double-stranded helix were also termed extruded helical single strands and described as Structural Classification of RNA (SCOR) motifs (6,15). As estimated by the recent study of Ellis and Jones (16), the flexibility in the protein binding sites is not significant and should allow structural prediction of protein–RNA interaction. In our recent work (17), we investigated the protein binding pockets that accommodate extruded nucleotides. We observed that most of the protein interacting nucleotides are part of a consecutive fragment of at least two nucleotides, whose rings have significant interactions with the protein. Many such pairs were observed to share the same protein binding cavity and >30% of these pairs are π-stacked. We showed that the classification of the nucleotide and dinucleotide binding sites reveals similarities in patterns important in protein–RNA recognition. We further showed that searching for known binding patterns on the surface of a target protein allows the prediction of its dinucleotide binding sites with a high success rate (17).
Here, we present the RNA binding site Data Base (RsiteDB), which details and classifies the nucleotide and dinucleotide binding pockets from all known protein–RNA complexes. The database contains the 3D physico-chemical patterns that describe the main types of the interactions. The on-line search engine of RsiteDB allows to search these patterns in a query protein. This predicts the binding sites and the binding modes of RNA dinucleotide. The on-line structural search against the entire data set of 3D patterns takes only several minutes and provides an atomic level prediction of protein–RNA interactions.
RsiteDB overview
As illustrated in Figure 1, RsiteDB contains the nucleotide binding sites extracted from all known protein–RNA complexes. These binding sites are classified into clusters according to the spatial arrangement of the protein physico-chemical and geometrical properties. The created clusters provide a set of 3D consensus binding patterns, which represent the main types of protein–nucleotide interactions. This classification is useful both for the analysis of existing interactions and for prediction of unknown ones.
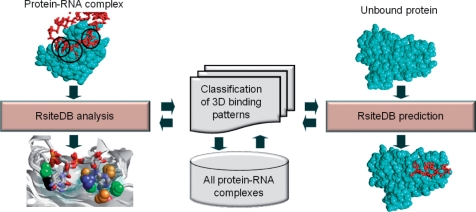
Figure 1.
RsiteDB overview. An overview of RsiteDB infrastructure and modes of operations. RsiteDB is based on the classification of the 3D binding patterns extracted from all protein–RNA complexes. There are two ways to query RsiteDB. First, given a protein–RNA complex, RsiteDB analyzes and classifies its protein–nucleotide interactions. Second, given an unbound protein structure, RsiteDB predicts the regions that can function as dinucleotide binding sites.
The database search engine allows data retrieval or 3D searches with a query structure. The first option allows the analysis of existing complexes, we refer to it as RsiteDB analysis subsequently. This retrieves the properties and the similarities of nucleotide and dinucleotide binding sites stored in the database. The second option allows the prediction of novel interactions of unbound proteins. We refer to it as RsiteDB prediction. This is a different type of search algorithm which performs an online structural search of the query protein against the database of 3D-consensus binding patterns. The search algorithm is based on an efficient Geometric Hashing algorithm (18), which allows a simultaneous comparison to all of the database patterns (17). Regions that are structurally and physico-chemically similar to any of these patterns are predicted to serve as binding sites. The RNA nucleotides, bound to the top ranking patterns from the database, predict the binding modes of nucleotides to the query protein.
Below we detail the information provided by RsiteDB and the different ways to query it. The sections are organized according to the screens presented to the user at the different stages of the analysis (see Figures 2–4).
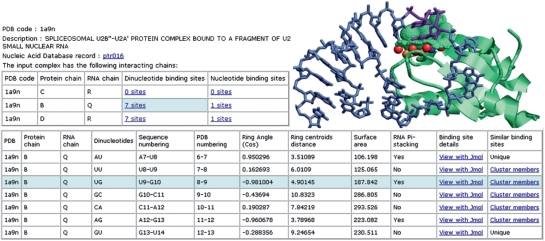
Figure 2.
Analysis of dinucleotide binding sites. The top left figure illustrates the basic analysis of the input protein–RNA complex. For each pair of interacting chains it presents the number of nucleotide and dinucleotide binding sites. Making the selection marked in light blue allows the user to explore the table of dinucleotide binding sites presented at the bottom. The top right figure illustrates one of the dinucleotide binding sites, which is marked in light blue. The extruded RNA nucleotides are purple sticks and the surface of the protein binding pocket is represented by green dots. The protein pseudocenters are represented as balls. Hydrogen bond donors are—blue, acceptors—red, donors/acceptors—green, hydrophobic aliphatic—orange and aromatic—white/gray.
RsiteDB analysis
Given the structure of a protein–RNA complex (specified by its PDB code), RsiteDB details its interacting protein-RNA chains. For each pair of chains, RsiteDB details the number of atomic contacts, which are defined by atoms within a distance of 5 Å. A pair of protein–RNA chains is considered to be interacting if there are at least 10 atomic contacts between them. We further analyze the protein interacting nucleotides and dinucleotides, their geometries and the properties of the corresponding protein binding sites. We define a nucleotide binding site by the protein Connolly solvent accessible surface area (19) within 2 Å from the surface of the RNA base. Nucleotides with a protein binding site area larger than 3 Å2 are defined as protein interacting. Given a pair of extruded consecutive nucleotides that interact with the protein, a dinucleotide binding site is defined by a pair of corresponding nucleotide binding sites.
Figure 1 presents the details provided by RsiteDB and illustrates one binding pocket accommodating a pair of consecutive π-stacked nucleotides. RsiteDB presents such parameters as the distance and angle between consecutive nucleotides as well as the binding site surface area. RsiteDB considers the protein binding sites represented by their surfaces and the physico-chemical properties termed pseudocenters (20). These are points in 3D space extracted from the protein amino acids that represent groups of atoms according to the interactions in which they may participate: hydrogen-bond donor (DON), hydrogen-bond acceptor (ACC), mixed donor/acceptor (DAC), hydrophobic aliphatic (ALI) and aromatic contacts (PI).
RsiteDB classification
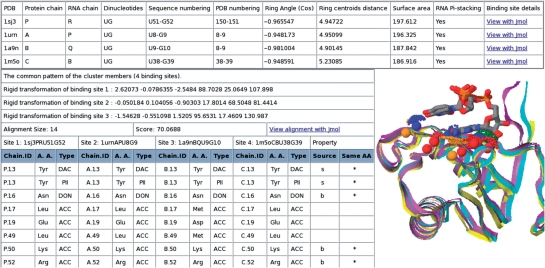
Each binding site from a known protein–RNA complex is either assigned to a cluster of similar binding sites or described as unique. Members of the same cluster share a similar spatial arrangement of pseudocenters, which we term a 3D consensus pattern. For each cluster, RsiteDB details its members and their multiple binding site alignment. The multiple alignment of nucleotide and dinucleotide binding sites is performed with the MultiBind and RnaBind methods, respectively (17). Figure 3 illustrates the analysis of the 3D consensus pattern described by the pseudocenters matched by the alignment. For each matched pseudocenter, we present its property as well as the details of the amino acid that originated it. As illustrated in Figure 3, RsiteDB provides a Jmol visualization of the multiple alignment and of the common pattern shared by the cluster members.
Figure 3.
RsiteDB classification. An example of a cluster of dinucleotide binding sites. RsiteDB details the properties of the binding sites in the cluster (top table) and provides the transformations that can align them in 3D space. The bottom right table details the matched pseudocenters of the common pattern. Each binding site in a cluster is described by its PDB code, chain identifies and nucleotide identities (e.g. 1sj3PRU51G52). It has three columns which provide the following details of its matched pseudocenters: (i) chain identifier and residue number; (ii) residue type and (iii) pseudocenter type. Although the pseudocenters are not required to have the same amino acid identity or origin (backbone or side chain), we indicate the conservation of these (* or b/s, respectively). The RNA dinucleotides are represented as sticks, colored by their atoms.
The classification of RsiteDB is unique due to several reasons. First, it accounts for the spatial physico-chemical properties of dinucleotide binding sites. Second, it is based on a classification methodology, which performs multiple binding sites alignment and validates the spatial superimposition of the cluster members. This overcomes the problem that objects similar in pairs may not be similar as a whole group. Specifically, by using multiple alignment, we assess the quality of the constructed 3D consensus pattern, which is required to be shared by all cluster members and to constitute at least 30% of each binding site (17).
The classification algorithm was applied to a non-redundant data set of protein–RNA complexes, which is provided at the RsiteDB web site. This data set was constructed by considering all high-resolution X-Ray structures of protein–RNA complexes [NDB release May 2008 (21), resolution better than 3 Å]. We extracted all pairs of interacting chains and removed protein and RNA chains with sequence identity above 25% and 60% in both chains, respectively. The classification of this non-redundant data set created 61 clusters of binding sites with more than one member. Approximately 44% of these clusters involve proteins with different sequences [<25% similarity and different Pfam annotations (25)]. Complexes that were removed due to redundancy, were added at the later stages of classification, and were assigned to the cluster of the closest homologue that fulfills the classification requirement (i.e. the constructed 3D consensus pattern is at least 30% of each of the cluster members). Using this procedure, 60% of all the dinucleotide binding sites and 45% of the single nucleotide binding sites were assigned to a cluster with more than one member. The same procedure was applied to the available NMR structures, which are assigned to the created clusters and are analyzed by RsiteDB.
RsiteDB prediction
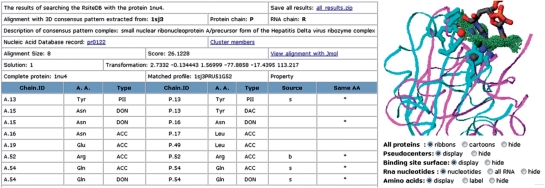
Here, we use the created clusters to predict the RNA binding sites that accommodate unpaired extruded nucleotides. Specifically, given a potentially unbound protein structure, we search its surface for regions similar to the 3D consensus binding patterns. These are defined by the above described clusters. Due to the low number of significant clusters of single nucleotide binding sites, currently only the dinucleotide patterns are used for the prediction. The search is performed with the RnaPred algorithm (17), which outputs a list of alignments to different protein regions that are recognized to contain some of the constructed 3D patterns. For each alignment, we detail the rigid transformation that can superimpose the pattern upon the protein in 3D space. We apply the transformation and provide a PDB complex of the solution which includes the query protein, the superimposed 3D patterns, its binding site surface and RNA dinucleotides. Figure 4 presents an example of output page which details the matched pseudocenters and visualizes the predicted complex. These results can be viewed on-line with Jmol. Alternatively, the user can download all of the alignments with the corresponding PDB files of their superimposition. We provide scripts for the Rasmol software, allowing an off-line visualization of the results.
Figure 4.
Prediction of dinucleotide binding sites. The results of searching RsiteDB with a query protein. The table presents the details of a 3D pattern. It describes the complex which initiated the cluster and was used for the pattern construction. Similarly to Figure 3, it details the pseudocenters matched by the alignment. These provide the prediction of the interactions of the query protein with the nucleotide bases. As shown in the right figure, RsiteDB visualizes the constructed complex. The query protein is cyan and the amino acids involved in the predicted interactions are represented by sticks. The protein which represents the pattern is magenta ribbons and its binding site surface is green dots. The pseudocenters and the RNA dinucleotides are colored as in Figure 1.
Using leave-one-out tests, the success rate of these predictions was estimated to be ∼75% (17). Interestingly, 32% of the correct predictions were made based on proteins with different sequences (<25% identity and no common Pfam domain) and could not be obtained based on recognition of sequence motifs. The main contribution of our knowledge-based predictions is that they describe the protein physico-chemical patterns that may be involved in interactions and predict the spatial orientation of the RNA nucleotides in the protein binding site independent of the protein overall sequences or folds.
PERFORMANCE AND AVAILABILITY
All of the files that describe the classification and its data sets are provided at the website. The classification is performed off-line and the data retrieval is immediate. The prediction algorithm, which screens the protein of interest against the database of 3D consensus patterns, is extremely fast with an average running time of 3 min. In the case of longer running times, caused by the large size of the query protein or the server overload, the user can provide an email to which a link to the output page will be sent. The visualization of the results is based on Jmol, which requires a web browser that supports Java applets.
FUNDING
Clore PhD fellowship (A.S.-P.); Israel Science Foundation (281/05 to H.J.W.); National Institute of Allergy and Infectious Diseases (NIAID); National Institute of Health (1UC1AI067231); Binational US-Israel Science Foundation (BSF); Hermann Minkowski-Minerva Center for Geometry at TAU; National Cancer Institute; National Institutes of Health (contract NOI-CO-12400); Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding for open access charges: SAIC-Frederick, Inc.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Maxim Shatsky for his contribution to the center-star classification algorithm development. We also thank Oranit Dror for contribution of code to this project. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
REFERENCES
- 1.Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 2.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murzin A, Brenner S, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: Structural Classification of RNA, Version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 8.Antson AA. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 2000;10:87–94. doi: 10.1016/s0959-440x(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 9.Jeong E, Chung IF, Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Inform. 2004;15:105–116. [PubMed] [Google Scholar]
- 10.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim OTP, Yura K, Go N. Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Res. 2006;34:6450–6460. doi: 10.1093/nar/gkl819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YC, Lim C. Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2008;36:e29. doi: 10.1093/nar/gkn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 15.Klosterman PS, Hendrix DK, Tamura M, Holbrook SR, Brenner SE. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis JJ, Jones S. Evaluating conformational changes in protein structures binding RNA. Proteins. 2007;4:1518–1526. doi: 10.1002/prot.21647. [DOI] [PubMed] [Google Scholar]
- 17.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson H. Prediction of interacting single-stranded RNA bases by protein binding patterns. J. Mol. Biol. 2008;379:299–316. doi: 10.1016/j.jmb.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamdan Y, Wolfson H. Tampa, Fl, USA: 1988. Geometric hashing: A general and efficient model-based recognition scheme. In; pp. 238–249. Proceedings of the IEEE International Conference on Computer Vision. [Google Scholar]
- 19.Connolly M. Analytical molecular surface calculation. J. Appl. Cryst. 1983;16:548–558. [Google Scholar]
- 20.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence or fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 2003;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]