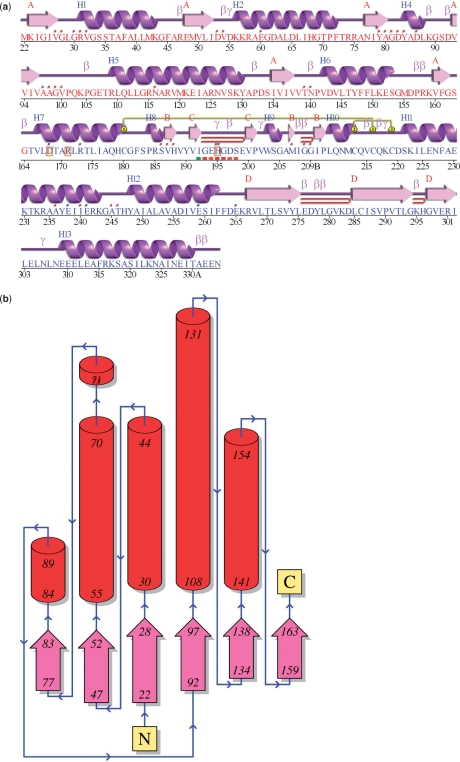
Figure 2.
Schematic diagrams from the PDBsum ‘Protein page’ for entry 1a5z: lactate dehydrogenase from Thermatoga maritima (16). (a) The ‘wiring diagram’ shows the protein's secondary structure elements (α-helices and β-sheets) together with various structural motifs such as β- and γ-turns, and β-hairpins. The yellow linking bars labelled 1 and 2 represent disulphide bonds. The single-letter amino acid codes showing the protein's sequence are coloured red or blue depending on whether they belong to CATH structural domain 1 or 2, respectively. Catalytic residues are indicated by a box surrounding the amino acid code. Red dots above the single-letter codes signify residues that interact with any bound ligand(s) while coloured lines underneath represent residues belonging to a PROSITE pattern, the redder the colour the more highly conserved the residue in the pattern. (b) Topology diagram of the first (i.e. red) structural domain in 1a5z. The diagram illustrates how the β-strands, represented by the large arrows, join up, side-by-side, to form the domain's central β-sheet. The diagram also shows the relative locations of the α-helices, here represented by the red cylinders. The small arrows indicate the directionality of the protein chain, from the N- to the C-terminus. The numbers within the secondary structural elements correspond to the residue numbering given in the PDB file.