Abstract
Recent advances in studies of Autism Spectrum Disorders (ASD) has uncovered many new candidate genes and continues to do so at an accelerated pace. To address the genetic complexity of ASD, we have developed AutDB (http://www.mindspec.org/autdb.html), a publicly available web-portal for on-going collection, manual annotation and visualization of genes linked to the disorder. We present a disease-driven database model in AutDB where all genes connected to ASD are collected and classified according to their genetic variation: candidates identified from genetic association studies, rare single gene mutations and genes linked to syndromic autism. Gene entries are richly annotated for their relevance to autism, along with an in-depth view of their molecular functions. The content of AutDB originates entirely from the published scientific literature and is organized to optimize its use by the research community. The main focus of this resource is to provide an up-to-date, annotated list of ASD candidate genes in the form of reference dataset for interrogating molecular mechanisms underlying the disorder. Our model for consolidated knowledge representation in genetically complex disorders could be replicated to study other such disorders.
INTRODUCTION
Autism (MIM 209850) is a broad-spectrum multifactorial condition with onset in the first years of life persisting throughout the lifetime (1). A triad of deficits in the areas of social communication, language development, repetitive activities and restricted range of interests define the core symptoms used in the diagnosis of autism (DSM IV, 1994). Autism Spectrum Disorders (ASD) is a commonly used term to cover the wide variations of autism. The dramatic rise in the prevalence of ASD in recent years is of major public concern (2,3).
A strong genetic component underlying ASD is firmly established from various lines of studies (4–7). The search for ‘causative’ gene(s) has resulted in >10 whole genome scans reporting numerous putative linkage regions for ASD susceptibility (8,9). Genetic association studies have identified many candidate genes for ASD (10–12); however, many candidates fail to replicate between studies and populations. In a minor proportion of cases, chromosomal aberrations have been identified (13). Recently, submicroscopic copy number variations (CNVs) were strongly associated with ASD (8,14,15). Additionally, ASD is consistently associated with a number of specific genetic disorders such as Fragile X syndrome amongst others (16,17). Single-gene mutations are also linked to rare cases of ASD (18,19). The high genetic heterogeneity of ASD poses an enormous challenge for understanding disease etiology.
We have developed an autism gene database, AutDB, for ongoing cataloguing of genes linked to the disorder. Our model for representing genetic knowledge encompasses collecting all types of genes including monogenic and risk-conferring candidates linked to ASD. We have implemented an integrated informatics approach to richly annotate the candidate genes for their relevance to autism, as well as an in-depth view of molecular functions. The focus of this resource is to provide up-to-date, annotated list of ASD candidate genes to serve as reference datasets to understand disease biology. To the best of our knowledge, this is the first example of an integrated gene database for a genetically complex disorder.
DISEASE-SPECIFIC KNOWLEDGE MODEL OF AutDB
AutDB was designed and developed as an integrated, disease-driven database model where both monogenic and small risk-conferring candidates associated with ASD are collected and annotated with diverse information (Figure 1). In this resource, ASD-related genes are classified into four categories including three genetic categories based on the type of genetic variation: (i) rAut: This category includes genes implicated in rare monogenic forms of ASD. The types of allelic variants within this class include rare polymorphisms and single-gene disruptions/mutations directly linked to ASD, together with sub-microscopic deletions/duplications (CNVs) encompassing single-genes specific for ASD. (ii) sAut: Genes implicated in syndromic forms of autism where a sub-population with a specific genetic syndrome develops autistic symptoms (iii) iAut: Small risk-conferring candidate genes with common polymorphisms identified from genetic association studies in idiopathic ASD, and (iv) fAut: Functional candidates relevant for ASD biology, not covered by any of the other genetic categories. However, it is possible that a gene can belong to more than one category depending on the mutation; a common variant conferring risk for developing idiopathic autism, while an inactivating mutation in the same gene placing it in higher risk-conferring categories. In such cases, all categories are used to annotate the genes. Both rAut and sAut categories represent monogenic forms of ASD, however, we made a distinct class for each based on the fact that rAut genes were identified in the course of screening for genetic variants in individuals diagnosed with ASD. In contrast, ASD is diagnosed secondary to the main clinical features of the specific genetic disorder within the syndromic forms. An additional category of functional candidates (fAut) extracted from reports linking a gene/protein to the biology of ASD, are also included in this resource and annotated accordingly.
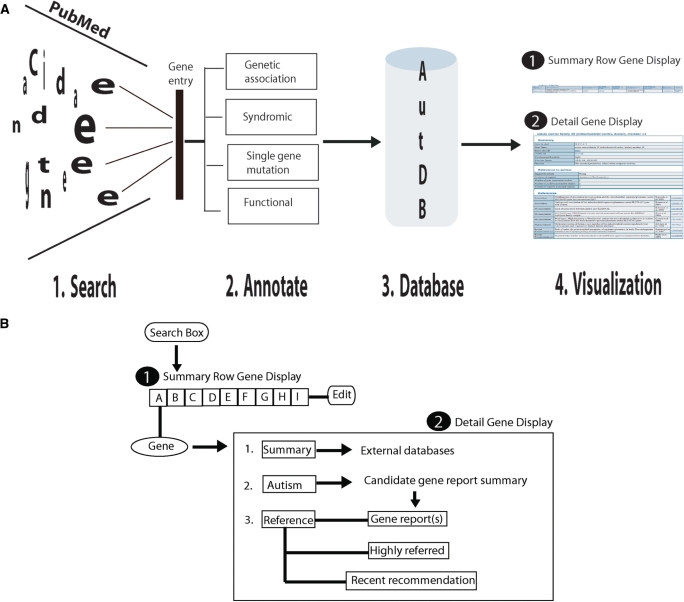
Figure 1.
(A) An integrated model for collection, annotation, storage and visualization of candidate genes for ASD. (1) Candidate genes for ASD are compiled and maintained from an exhaustive search of scientific literature in the PubMed database maintained at NCBI (2) ASD candidates are classified into four categories. (3) Entries are organized in the database for search and retrieval. (4) User-friendly display of candidate genes at two levels for display and easy access with links to various external databases. (B) Search and display of candidate genes in AutDB. A search box is used to retrieve ASD candidate genes catalogued in AutDB using various gene attributes. The search results are displayed at two levels. At level 1, the gene entry is displayed in the summary row format showing: (A) gene symbol, (B) gene name, (C) chromosomal location, (D) Genetic category, (E) GAD and (F) OMIM links where available, (G) Number of autism-specific studies, (H) primary PubMed reference and (I) detail/edit button. A level 2, the entry is further displayed at a detail level in three-tier format showing: (1) gene summary with links to external databases such as Entrez Gene and UniProt, (2) relevance for autism displaying candidate gene report summary and (3) references arranged under three sub-headings: references for the candidate gene reports, highly referred and recent studies of the candidate genes.
AutDB: THE AUTISM GENE DATABASE
The content of AutDB originates entirely from published scientific literature and is manually annotated by expert biologists. A comprehensive collection of ASD-related genes was initially compiled from an exhaustive search of the scientific literature from PubMed database at NCBI (http://www.ncbi.nih.gov/pubmed), followed by timed-searches to maintain an up-to-date resource (see Methods section for details). An AutDB entry is a candidate gene linked to ASD with all its attributes. The steps involved in the process for curation of an AutDB entry are schematically shown in Figure 1. First, all reports pertaining to a candidate gene are extracted, counted for the number of studies and collapsed under a single header representing the gene entry. Second, a multi-step annotation strategy is implemented to incorporate diverse molecular information about a candidate gene for assessment of its relevance for ASD. To enhance functional knowledge of a candidate gene, our annotation model also expands entries by drilling down to highly cited articles on candidate gene/protein, together with recently published articles to represent current knowledge of gene functions. This feature of AutDB provides functional information of an ASD candidate gene beyond the basic information available in large public databases. Finally, candidate genes are classified based on the type of genetic category according to AutDB knowledge model (described in the previous section). An example of an annotated AutDB gene is shown in supplementary Figure S1.
SEARCH AND DISPLAY OF AutDB DATA
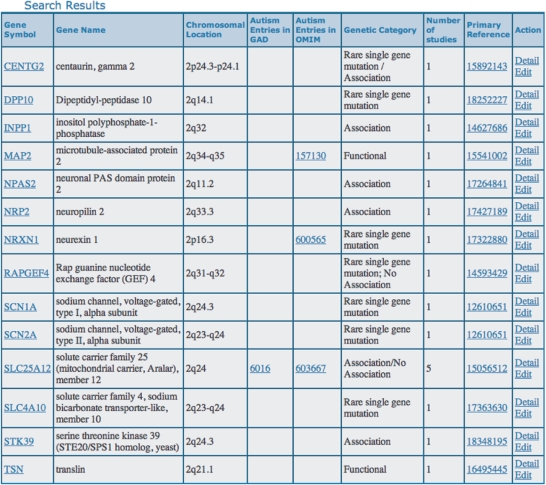
Genetic information in AutDB can be searched and displayed in several ways including complex Boolean queries. A representative search result of AutDB is shown in Figure 2. In this example, searching for ASD candidate genes on Chromosome 2 will retrieve a comprehensive list of ASD candidate genes in a tabular format. This list includes candidates reported from genetic association studies, together with rare single-gene mutations and functional candidates. Searching by gene name or gene symbol will retrieve a gene entry that can be displayed at two levels (Figure 1B). At the first level of display in the summary row format, each entry is annotated with gene symbol, gene name, chromosomal location, genetic category and GAD and OMIM links where available, total number of autism-specific studies used to create the entry in AutDB, together with a primary PubMed reference reporting the candidate gene. Each entry is further displayed at a detail level showing (i) gene summary with links to external databases such as Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez) and UniProt (http://www.uniprot.org/) to provide standardized generic information on candidate genes, (ii) relevance for autism showing autism-specific information describing the type of genetic variants linked to ASD and the studies reporting the gene and (iii) references arranged under three subheadings. First, references linking the gene to ASD are displayed. A primary reference in AutDB is defined as the first positive report linking the candidate gene to ASD. Next, highly referred studies on the candidate gene extracted using Google scholar based on number of citations are shown. Finally, references for recent studies pertaining to the candidate genes extracted from timed-search of PubMed are also provided. A flow-chart describing the search and display of an AutDB entry is shown in Figure 1B. Annotated list of ASD candidate genes can also be obtained from AutDB to serve as primer for building customized reference datasets. These reference datasets provide global attributes of ASD-linked genes and define an entry point for further enhancement and optimization based on specific research requirements.
Figure 2.
Online display of AutDB search results. (A) Searching for candidate genes on Chromosome 2 retrieved an annotated list of ASD candidate genes in Chromosome 2.
DISCUSSION
AutDB has been developed to provide current knowledge on candidate genes linked to ASD and their relevance for the disorder. We have adopted several novel strategies to design the resource with the focus on gene functions. For example, each candidate gene is annotated with highly referred citations, together with recent recommendation to provide a panoramic view of gene function. In our view, AutDB annotation model can be applied to annotate protein entries in other databases. Compared to other disease-specific databases such as AlzGene (http://www.alzforum.org/res/com/gen/alzgene/), which collects only candidates identified from genetic association studies, our integrative model includes all types of genetic variations implicated in ASD. A recent search of OMIM database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM) for genes linked to autism retrieved ∼60 entries (August, 2008), compared to 133 entries in AutDB as of this writing. A recent review article lists 26 candidate genes (12), thereby affirming that without a systematic and sustained effort it is difficult to keep up with the rapidly evolving field of autism biology. The first rare single gene mutation in ASD was reported in 2003 (18), however, in the last two years with the application of genomic technologies, a steady stream of single gene mutations in ASD is reported in the scientific literature (15,20). Consequently, the balance is shifting between reported single-gene mutations and candidates identified from genetic association studies. The current trend in autism genetics validates the integrative model in AutDB.
Our major focus is to provide a filtered, annotated reference set of ASD-linked genes to the research community for immediate application in the areas of (i) building predictive disease models using bioinformatics (manuscript in preparation) and (ii) systems-level analysis of various molecular data to gain insight into the etiology of ASD.
PERSPECTIVE
Autism, a rare encounter in mid last century, is becoming increasingly prevalent in the last two decades. Accumulating evidence strongly point towards many genetic causes can contribute to the development of ASD. In practical terms, the number of articles reporting putative candidate loci, as well as high throughput array-based studies reporting many loci in a single publication are rapidly accumulating. We have created AutDB to bridge the gap between the vast amount of information embedded in the scientific literature and consolidated knowledge representation of ASD for molecular analysis. Lastly, the dynamic nature of this database provides a window into the current state of research to aid in the early diagnostics and therapeutics for individuals with autism.
METHOD
Design of an integrated platform
AutDB is a portal developed in JAVA on the J2EE platform on Linux with an Relational Database Management System (RDBMS) backend as its repository. The portal for AutDB is designed to be extensible where newer modules could be incorporated with relative ease by configuration. The application is deployed as a webapps in the Tomcat Application server connecting to the RDBMS. Connection pooling is provided by the Application Server, which greatly decreases the load on the system and enhances the performance. It also connects to the NLM database with the help of their DTDs and collects relevant information from the NLM databases for the end user. Apart from batch mass updates and new additions through database load programs, AutDB also provides interfaces for online updates restricted by user roles. Most importantly, AutDB provides interfaces within the Autism research community for co-operative, moderated annotations and curations for deeply annotated genes or gene sets. A search engine is provided on different attribute names on their applicable data sets to build queries at varying degrees of complexity.
Development of AutDB
Collection, storage and categorization of candidate genes
To collect ASD candidate genes for the first release of AutDB, we performed a comprehensive search of all articles in the PubMed database maintained at NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed). The search terms included ‘gene’ AND (‘autism’ OR ‘autistic’) restricted to the titles and abstracts of the publications for retrieval. Furthermore, candidate genes listed in review articles on molecular genetics of ASD, along with cross-references therein, were mapped and added (if new) to our candidate gene list from PubMed searches to compile the most exhaustive gene set. After the first release, starting from June 2006, a daily semi automated search of the PubMed with the same keywords is implemented to maintain an up-to-date resource of all candidate genes linked to ASD. Additionally, relevant journal articles in the fields of genetics, neurobiology, and psychiatry are screened on a regular basis to enrich the resource.
To select the primary reference for a candidate gene, the first positive report linking the gene to (ASD) was searched in PubMed. However, to select the primary reference for a ‘Syndromic autism’ gene where a large number of reports are published connecting the syndrome to autism, we adopted the following steps: First, google scholar is used with the criteria [(autism OR autistic) AND ‘syndrome name’ AND ‘gene name’] to search for the most highly cited reports. Next, a primary reference is selected among these highly cited reports by reading the article (see Supplementary Table 1).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: MindSpec Inc., a non-profit organization dedicated to autism research.
Conflict of interest statement: MindSpec and Sharmila Banerjee-Basu hold the license for AutDB.
REFERENCES
- 1.Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 2.Van Naarden Braun K, Pettygrove S, Daniels J, Miller L, Nicholas J, Baio J, Schieve L, Kirby RS, Washington A, Brocksen S, et al. Evaluation of a methodology for a collaborative multiple source surveillance network for autism spectrum disorders–Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2002. MMWR Surveill. Summ. 2007;56:29–40. [PubMed] [Google Scholar]
- 3.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J. Clin. Psychiatry. 2005;66(Suppl. 10):3–8. [PubMed] [Google Scholar]
- 4.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 5.Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M. A broader phenotype of autism: the clinical spectrum in twins. J. Child Psychol. Psychiatry. 1996;37:785–801. doi: 10.1111/j.1469-7610.1996.tb01475.x. [DOI] [PubMed] [Google Scholar]
- 6.Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat. Rev. Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children. JAMA. 2001;285:3093–3099. doi: 10.1001/jama.285.24.3093. [DOI] [PubMed] [Google Scholar]
- 8.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 10.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int. J. Dev. Neurosci. 2007;25:69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol. Psychiatry. 2006;1:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 14.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J. Dev. Behav. Pediatr. 2001;22:409–417. doi: 10.1097/00004703-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Cohen D, Pichard N, Tordjman S, Baumann C, Burglen L, Excoffier E, Lazar G, Mazet P, Pinquier C, Verloes A, et al. Specific genetic disorders and autism: clinical contribution towards their identification. J. Autism Dev. Disord. 2005;35:103–116. doi: 10.1007/s10803-004-1038-2. [DOI] [PubMed] [Google Scholar]
- 18.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]