Abstract
OryGenesDB (http://orygenesdb.cirad.fr/index.html) is a database developed for rice reverse genetics. OryGenesDB contains FSTs (flanking sequence tags) of various mutagens and functional genomics data, collected from both international insertion collections and the literature. The current release of OryGenesDB contains 171 000 FSTs, and annotations divided among 10 specific categories, totaling 78 annotation layers. Several additional tools have been added to the main interface; these tools enable the user to retrieve FSTs and design probes to analyze insertion lines. The major innovation of OryGenesDB 2008, besides updating the data and tools, is a new tool, Orylink, which was developed to speed up rice functional genomics by taking advantage of the resources developed in two related databases, Oryza Tag Line and GreenPhylDB. Orylink was designed to field complex queries across these three databases and store both the queries and their results in an intuitive manner. Orylink offers a simple and powerful virtual workbench for functional genomics. Alternatively, the Web services developed for Orylink can be used independently of its Web interface, increasing the interoperability between these different bioinformatics applications.
INTRODUCTION
Reverse genetics is a powerful way to discover the biological role of thousands of genes whose functions are currently unknown. Currently, large-scale random insertion mutagenesis is one of the most powerful gene inactivation methods for reverse genetics (1). Several laboratories around the world have developed T-DNA insertion libraries with various constructs [see (1) for a review]. The systematic sequencing of DNA flanking the mutagens is a simple way to identify disrupted lines for virtually any rice gene. OryGenesDB is the most complete open-access database for rice reverse genetics and compiles large-scale insertion mutagenesis data from several laboratories. OryGenesDB is now a standard resource for rice reverse genetics and has greatly expanded since its first release in 2006 (2).
Several additional databases of interest for functional genomics have recently been released, including Oryza Tag Line (3), which contains phenotypic descriptions of the Genoplante insertion lines, and GreenPhylDB (4), a comprehensive platform designed to facilitate comparative functional genomics between the Oryza sativa and Arabidopsis thaliana genomes. In order to facilitate user navigation between these three databases, and as a first step towards database interoperability for rice functional genomics, we developed a specific tool called Orylink. Orylink is a personalized integrated system for functional genomics analysis and is fully integrated into OryGenesDB. This modular system was developed to carry out complex queries across these three databases using different entry points, including TIGR (5), InterPro (6), KEGG id (7) or keywords. We developed Web services for interoperability between the OryGenesDB, Oryza Tag Line and GreenPhylDB databases, which can also be used as a stand-alone system to query across these three databases using BioMOBY (8,9). Using Orylink and its associated Web services, a user can retrieve and store information, including FSTs, mutant phenotypes and A. thaliana orthologs, across these three databases on a project basis. A classical query example is to start from a candidate rice gene, look for the A. thaliana orthologs (GreenPhylDB), identify the corresponding rice mutant insertion lines (OryGenesDB), and finally check if any of these lines has an already characterized phenotype (Oryza Tag Line). Orylink can field such classical queries, as well as more complex queries, across these three databases and others. Here, we present a major expansion of OryGenesDB that includes Orylink as well as some new tools and data.
RESULTS
Expansion of OryGenesDB
FST
OryGenesDB is specifically dedicated to rice reverse genetics and attempts to map FSTs from international laboratories. Since the release of OryGenesDB in 2006, when around 45 000 FSTs were available, more than 125 000 additional FSTs have been added, for a current total of 171 000 FSTs from 10 laboratories (Table 1). OryGenesDB is regularly updated, with updates including genome annotations [version 5 (10) and 7 (11) of the TIGR (O. sativa) and TAIR (A. thaliana) annotations, respectively] and the GMOD browser, which offers new user friendly interfaces through AJAX technologies (12). A new interface with a tab-like organization of sub-menus was developed to simplify navigation across OryGenesDB tools.
Table 1.
Rice insertion resources integrated in OryGenesDB
| Institution | Mutagen | No. of flanking sequences | No. of mapped sequences |
|---|---|---|---|
| CerealGene Tags, European Union | Ac/Ds | 1380 | 1380 |
| CIRAD, Genoscope | Tos17 | 13 745 | 13 622 |
| CIRAD-INRA-IRD-CNRS, Genoplante | T-DNA | 14 137 | 13 384 |
| CSIRO | T-DNA | 787 | 787 |
| National Center of Plant Gene Research | T-DNA | 16 158 | 15 807 |
| National Institute of Agrobiological Sciences | Tos17 | 18 024 | 17 955 |
| Plant Functional Genomics Laboratory | T-DNA | 80 006 | 78 709 |
| PMBBRC | Ac/Ds | 1072 | 1044 |
| Taiwan Rice Insertional Mutant Program | T-DNA | 11 799 | 11 754 |
| University of California at Davis | Ac/Ds | 13 922 | 12 927 |
| Total | 171 030 | 167 369 |
When the phenotypic characterization of 30 000 T-DNA enhancer trap lines from the Genoplante library was recently completed, the data were stored in Oryza Tag Line (OTL) (3). The corresponding FSTs are stored in OryGenesDB, and we cross-linked OTL lines and the corresponding FSTs in OryGenesDB. The direct link to OTL appears in the pop-up window to allow a direct observation of the line phenotype, if any (3). Cross links are also available through Orylink (see below).
Automatic design of primers for FST validation
A new tool, called Primer blaster, was designed to test the specificity of any primer pairs on the rice genome. The user can paste or download primer sequences as multifasta files, with the reverse and forward primers in the same order in the two files. During the next step, the database, usually pseudochromosomes, is selected, and the expected amplicon size is fixed. After the query is submitted, a table is displayed that includes all of the primers tested with the primer name, the chromosome, the primer's position, the amplicon size and the number of hits for the forward and reverse primers on the rice genome, and their status is shifted to ‘ok’ if the primer pair is really specific.
Designing probes to analyze insertion lines is a very repetitive and common task. Therefore, we developed the ‘Primer Designer’ tool to help users develop probes for Southern blot. Users input either the candidate FST, plant name, a sequence size from FST or alternatively a sequence size range, and the restriction enzyme to be used for the Southern blot. Primer designer will then search for a probe around the FST, according to the user's search parameters, that do not contain the chosen restriction site. The probe can be then used to check if there is a rearrangement at the expected locus and also to identify the given FST as homozygous or heterozygous.
New sub-databases
OryGenesDB is not only a repository for FSTs but was also designed to store more specialized data related to functional genomics. Hence, several sub-databases, including Archipelago, were developed to store information on more 2500 genes of the rice defense arsenal (13) that were obtained from more than 70 publications. Similarly, extensive QTL cataloguing for Rice Blast resistance identified 85 blast resistance and around 350 QTL that were mapped on the rice genome and could be incorporated into OryGenesDB (14). These sub-databases are now accessible as specific categories in the Genome Browser tracks through ‘Resistance Gene Analogues’ and ‘Rice Defense genes’ for Archipelago and ‘Blast QTL and R-Genes’ for the Meta-QTL analysis.
New annotation layers
A total of 10 specific categories are accessible in OryGenesDB, totaling 78 annotation layers. For instance, the ‘Oryza Map Alignment Project’ (OMAP) aims to construct and align BAC/STC-based physical maps of 11 wild and one cultivated rice species of the c.v. Nipponbare, to exploit the potential of the wild species of the genus Oryza for breeding cultivated rice cultivars (15). In order to provide simple and fast access to this resource, all STC identified in the OMAP project were mapped to the Nipponbare pseudomolecule and are visible as a supplemental layer of annotation.
Extended functionalities
Orylink: comparative functional genomics across databases in a ‘click and view’ manner
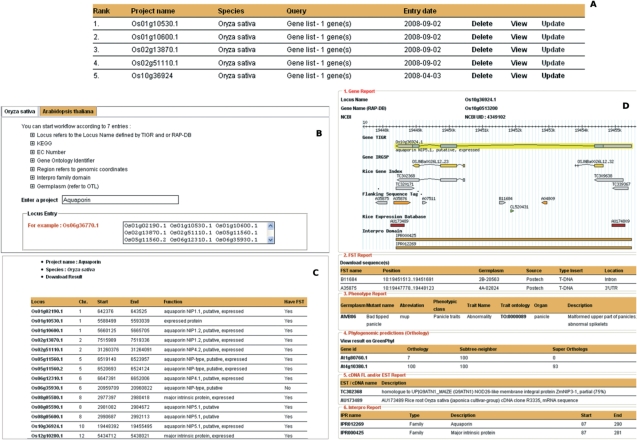
For the user's point of view, Orylink is like a virtual workbench that helps biologists retrieve all kind of information linked with the gene locus. Figure 1 shows an example of a biological query process.
Figure 1.
Data search process using Orylink. This figure illustrates the various steps to initiate a user query. Starting with the project framework (A), users can create new projects (B) and display the results (C and D) after providing a login and a password. (A) Project management interface. (B) Project creation interface. (C) A synthetic result obtained for the execution of a given workflow and (D) gene report for the corresponding locus name ‘0s10g36924.1.’ In this view, the data are organized into broad categories like gene, phenotype and phylogenomic predictions reports. Cross-references are built into all the data to link the summary data to their original sources.
Orylink is available through the tool menu of the main OryGenesDB tool bar (16). Starting with the project framework (Figure 1A), users start a new project (e.g. Aquaporin) (Figure 1B) after providing a login and password. Each project is focused on one species, either O. sativa or A. thaliana. Biologists can build their own queries according to seven different data types [TIGR ID (5), InterPro ID (6), KEGG ID (7), Enzyme Commission number (17), Gene Ontology ID (18), Germplasm ID (3) and genomic location]. In this case, a list of loci is submitted. Figure 1C shows the results of the Aquaporin project. In this synthetic view, 14 genes are retrieved. Users can quickly observe the locations of the genes and their annotations and whether the genes have either KO lines (identified in the ‘Have FST’ column) or reporter gene expressions and phenotypes (identified in the ‘Have expression’ and ‘Have phenotype’ columns, respectively). The column ‘supported by evidence’ represents the presence of cDNA and EST for the locus in question. The results can also be downloaded in the Excel TM file format. By clicking on the locus entry, users can display detailed results (Figure 1D). For these detailed results, Orylink first provides a GBrowse image of the genomic location corresponding to the locus entry. It lists all FSTs that disrupt the gene, with details of their origins (column source) and features (e.g. location, orientation and type of insert). It provides numerous features of the phenotype observations extracted from Oryza Tag Line, for example, the phenotype name with its description. Also, the phenotypic class joined with the trait ontology ID is displayed. Orylink extracts corresponding A. thaliana orthologs from GreenPhylDB. This information can be an open area in comparative genomics and may provide a bridge between A. thaliana and O. sativa functional genomics data. EST and cDNA features are displayed with their annotations to enrich some putative gene functions. Finally, a list of InterPro names is provided to identify all protein domain families.
One of the most important benefits of Web services is that access to original data sources guarantees up-to-date information. Another benefit is the ability to launch massive queries recursively with free access. Moreover, bioinformaticians can easily chain Web services with a minimum of programming (see Materials and Methods section of Supplementary data for the tools we used).
CONCLUSIONS AND FUTURE DIRECTIONS
OryGenesDB is now not only the most popular and complete database for rice reverse genetics, but it is also a user-oriented web application that automates and organizes Web queries in rice functional genomics. Biologists can greatly reduce the time wasted assembling data from heterogeneous data sources. In addition to complex queries across databases, OryGenesDB offers a simple way to store and organize hypotheses as projects through Orylink. Users can run their queries and store the output but also update the results as the source databases are continuously independently updated. OryGenesDB offer a way to accelerate the manual process of integrating and compiling data from heterogeneous sources. As a whole, this database is guided by the biologists' need for automation and integration. In the future, new workflows like the Nottingham Arabidopsis Stock Centre (NASC) (19) and the Munich Information center for Protein Sequences (MIPS) (20) will be developed or integrated into Orylink. Depending on the Web services available, new queries will be implemented, for instance, to assign rice gene functions using the available compiled gene-oriented literature on A. thaliana genes. Starting from a gene of interest in rice, an Orylink user will soon be able to find the corresponding A. thaliana genes and identify the putative functions of the rice gene using TAIR data. This functionality will greatly help any biologist with interest in the comparative functional genomics between rice and A. thaliana, the two plant models. Last, the integration of genomics data in OryGenesDB will continue, including new FSTs and new annotation layers. Users are also encouraged to submit proposals for interface modifications and data of interest that they want to integrate into OryGenesDB at orygenesdb@cirad.fr.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Funding for the open access charge: Centre de coopération internationale en recherche agronomique pour le développement.
ACKNOWLEDGEMENTS
The authors thank Emmanuel Guiderdoni and Manuel Ruiz for their support.
REFERENCES
- 1.Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, et al. Rice mutant resources for gene discovery. Plant Mol. Biol. 2004;54:325–334. doi: 10.1023/B:PLAN.0000036368.74758.66. [DOI] [PubMed] [Google Scholar]
- 2.Droc G, Ruiz M, Larmande P, Pereira A, Piffanelli P, Morel JB, Dievart A, Courtois B, Guiderdoni E, Perin C. OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res. 2006;34:D736–D740. doi: 10.1093/nar/gkj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larmande P, Gay C, Lorieux M, Perin C, Bouniol M, Droc G, Sallaud C, Perez P, Barnola I, Biderre-Petit C, et al. Oryza Tag Line, a phenotypic mutant database for the Genoplante rice insertion line library. Nucleic Acids Res. 2008;36:D1022–D1027. doi: 10.1093/nar/gkm762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte MG, Gaillard S, Lanau N, Rouard M, Perin C. GreenPhylDB: a database for plant comparative genomics. Nucleic Acids Res. 2008;36:991–998. doi: 10.1093/nar/gkm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Quackenbush J. Using the TIGR gene index databases for biological discovery. Curr. Protoc. Bioinformatics. 2003;Chapter 1 doi: 10.1002/0471250953.bi0106s03. Unit 1.6. [DOI] [PubMed] [Google Scholar]
- 6.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson MD, Senger M, Kawas E, Bruskiewich R, Gouzy J, Noirot C, Bardou P, Ng A, Haase D, Saiz Ede A, et al. Interoperability with Moby 1.0—it's better than sharing your toothbrush. Brief Bioinform. 2008;9:220–231. doi: 10.1093/bib/bbn003. [DOI] [PubMed] [Google Scholar]
- 10.TIGR rice genome annotations version 5. ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_5.0/(10 October 2008, date last accessed). [Google Scholar]
- 11.TAIR. Arabidopsis thaliana genome annotations version 7. ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/(7 November 2008, date last accessed).
- 12.GMOD. web site: http://www.gmod.org (17 November 2008, date last accessed).
- 13.Vergne E, Ballini E, Droc G, Tharreau D, Notteghem JL, Morel JB. ARCHIPELAGO: a dedicated resource for exploiting past, present, and future genomic data on disease resistance regulation in rice. Mol. Plant Microbe Interact. 2008;21:869–878. doi: 10.1094/MPMI-21-7-0869. [DOI] [PubMed] [Google Scholar]
- 14.Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 2008;21:859–868. doi: 10.1094/MPMI-21-7-0859. [DOI] [PubMed] [Google Scholar]
- 15.Wing RA, Ammiraju JS, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, et al. The oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- 16.OryLink home page. http://orygenesdb.cirad.fr/login.html (23 October 2008, date last accessed).
- 17.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blake JA, Harris MA. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr. Protoc. Bioinformatics. 2008;Chapter 7 doi: 10.1002/0471250953.bi0702s23. Unit 7.2. [DOI] [PubMed] [Google Scholar]
- 19.Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiol. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spannagl M, Haberer G, Ernst R, Schoof H, Mayer KF. MIPS Plant Genome Information Resources. Methods Mol. Biol. 2007;406:137–160. doi: 10.1007/978-1-59745-535-0_6. [DOI] [PubMed] [Google Scholar]