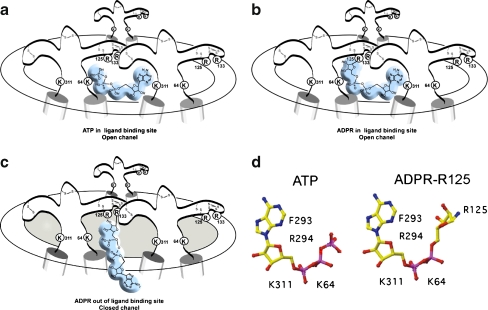
Fig. 5.
Model for the activation of P2X7 by ADP-ribosylation at R125. a Schematic diagram of the trimeric P2X7 receptor complex in open conformation following binding of ATP at the interface of two adjacent subunits. b ADP-ribosylation at R125 places the attached ADP-ribose in the ligand-binding site, inducing the open conformation. c ADP-ribosylation at residue R133 places the attached ADP-ribose out of reach of the binding site. d Schematic diagrams of the soluble ligand ATP and of ADP-ribose in covalent linkage to R125. Residues K64 and K311 may interact with the negatively charged phosphate groups, residues F293 and R294 with the adenine–ribose moiety [6, 76]