Abstract
The P2RX7 gene is highly polymorphic, and many single nucleotide polymorphisms (SNPs) underlie the wide variation observed in P2X7 receptor responses. We review the discovery of those non-synonymous SNPs that affect receptor function and compare their frequencies in different ethnic populations. Analysis of pairwise linkage disequilibrium (LD) predicts a limited range of haplotypes. The strong LD between certain functional SNPs provides insight into published studies of the association between SNPs and human disease.
Keywords: P2X7 receptor, Genetics, Single nucleotide polymorphisms, Linkage disequilibrium
Introduction
Major advances have been made in our understanding of the role the P2X7 receptor (P2X7R) has to play in human disease. Publication of the human genome sequence in 2004 [1] and the completion of the International HapMap Project in 2005 [2] has been followed by large genome-wide association studies that have found linkage for single nucleotide polymorphisms (SNPs) to human disease. It is generally assumed that SNPs identified through genetic association studies are excellent candidates for disease-causing variants. However, firm evidence for the significance of these candidates requires demonstration of their functional effect on the target protein. A potential complication of this approach comes from the existence of linkage disequilibrium within the genome such that the candidate gene may be co-inherited with an adjoining polymorphism that is the actual disease-causing variant. It is therefore timely to examine the pattern of inheritance of combinations of SNPs within the P2RX7 gene to define linkage disequilibrium between SNPs that have functional effects and to determine haplotype block structure of those SNPs that interact to modulate P2X7R function.
Recent advances in genetics
SNPs are the most common genetic variation in the human genome and there are currently over 14 million documented in dbSNP (www.ncbi.nlm.nih.gov/SNP; build 129), the public repository for genetic variation. Successful identification of disease-causing variations in genes before the completion of the Human Genome Project involved the use of linkage studies of large families with multiple affected members. These studies have been successful in identifying genes responsible for a number of diseases including BRCA1 and BRCA2 predisposing to breast cancer [3] and APC for familial adenomatous polyposis [4] although it has not been possible to identify susceptibility genes in the majority of complex diseases. It is hoped that with the availability of dense genetic maps of SNPs it will be possible to identify genes that contribute to susceptibility to complex common diseases using large association studies, and this approach has defined valuable loci for further study in bipolar disorder, rheumatoid arthritis, coronary artery disease, Crohn’s disease, and types 1 and 2 diabetes mellitus [5].
SNPs in P2RX7
The P2RX7 gene spans 53 kb with 13 exons and has been mapped to the 12q24.31 chromosomal region [6]. The P2RX7 gene is highly polymorphic and there are 32 non-synonymous, amino acid-altering SNPs currently documented in dbSNP (www.ncbi.nlm.nih.gov/SNP). The first to be functionally described was 1513A>C that changes glutamic acid to alanine at residue 496 (Glu496 to Ala) in the carboxyl terminus of the receptor and has an allele frequency of 0.14–0.18 in normal Caucasian populations [7]. The Glu496 to Ala polymorphism confers an unusual property upon the receptor in native or transfected cells since adenosine triphosphate (ATP)-induced ethidium uptake is abolished, but the immediate ATP-induced opening of the cation-selective channel is unaffected [8].
The second SNP to be described was 1729T>A that changes isoleucine to asparagine at residue 568 (Ile568 to Asn) which lies in a trafficking motif within the carboxyl tail of the receptor [9, 10]. This amino acid substitution prevents normal trafficking and cell surface expression of the receptor and transfection experiments have shown that P2X7R carrying the Ile568 to Asn mutation is non-functional. The 1729T>A SNP is rare in Caucasian populations with an allele frequency of 0.026 (Table 1). A third SNP, 946G>A, changes arginine to glutamine at residue 307 (Arg307 to Gln) in the ATP-binding pocket of the extracellular domain, and transfection experiments have shown that P2X7R carrying this mutation lacks channel and pore function although there is normal surface expression of the receptor [11]. A splice site mutation (g>t) at position +1 of the first intron causes a null allele in close to 1% of Caucasian subjects [12] due to nonsense-mediated mRNA decay. A fifth polymorphism (1096C>G) changes serine to threonine at amino acid 357 (Ser357 to Thr) and has an allele frequency of 0.075 in Caucasians. The Ser357 to Thr polymorphism produces a partial loss of P2X7R function which affects both channel and pore functions [13]. A sixth polymorphism (489C>T) changes amino acid 155 from histidine to tyrosine (His155 to Tyr) in the ectodomain [14] and has an allele frequency of 0.455 in Caucasians (Table 1). The His155 to Tyr polymorphism produces a gain-of-function effect on P2X7R pore function and calcium influx [14]. Finally, a seventh polymorphism has been described at 474G>A which reduces receptor function in native cells [15] although confirmatory transfection experiments are awaited to verify these data.
Table 1.
Minor allele frequencies of ten single nucleotide polymorphisms (SNPs) in the P2RX7 gene in an Australian Caucasian cohort
| SNP | Minor allele frequency |
|---|---|
| 151+1g>t | 0.008 |
| 474G>A | 0.015 |
| 489C>T | 0.455 |
| 853G>A | 0.023 |
| 946G>A | 0.016 |
| 1068G>A | 0.394 |
| 1096C>G | 0.075 |
| 1405A>G | 0.174 |
| 1513A>C | 0.185 |
| 1729T>A | 0.026 |
Genomic DNA was extracted from the peripheral blood of 1,755 normal Australian Caucasian subjects. Genotyping was performed using a Sequenom MassArray automated detection system at the Australian Genome Research Facility (Brisbane, Australia)
Re-sequencing of genes for the International HapMap Project (http://www.hapmap.org) has shown divergence in the polymorphic structure of the P2RX7 gene in different population samples. The HapMap data include nine non-synonymous SNPs in the P2RX7 gene: rs17525809 (253 bp), rs208294 (489 bp), rs7958311 (835 bp), rs7958316 (853 bp), rs1718119 (1068 bp), rs2230911 (1096 bp), rs2230912 (1405 bp), rs3751143 (1513 bp), and rs1653624 (1729 bp). These SNPs are all represented in CEPH (Utah Residents with Northern and Western European Ancestry) but of note is that rs7958311 (853 bp), rs2230912 (1405 bp), and rs1653624 (1729 bp) were not found in Japanese in Tokyo, Japan or Han Chinese in Beijing, China populations. Similarly, rs7958311 (853 bp) and rs1653624 (1729 bp) were not found in the Yoruba in Ibadan, Nigeria population. In addition, rs17525809 (253 bp) was not found in YRI and rs2230912 (1405 bp) minor allele frequency was very low (0.017) compared with the CEU population (Table 2). These differences in populations may reflect positive or negative selection pressures over long time periods by environmental and disease factors.
Table 2.
Minor allele frequencies of nine non-synonymous single nucleotide polymorphisms (SNPs) in P2RX7 from four populations described by the International HapMap Project (www.hapmap.org)
| SNP | Base pair P2RX7 | CEU (n = 120) | CHB (n = 90) | JPT (n = 90) | YRI (n = 120) |
|---|---|---|---|---|---|
| rs17525809 | 253 | 0.067 | 0.044 | 0.133 | – |
| rs208294 | 489 | 0.458 | 0.633 | 0.567 | 0.333 |
| rs7958311 | 835 | 0.183 | 0.400 | 0.344 | 0.233 |
| rs7958316 | 853 | 0.008 | – | – | – |
| rs1718119 | 1,068 | 0.492 | 0.122 | 0.211 | 0.517 |
| rs2230911 | 1,096 | 0.100 | 0.244 | 0.133 | 0.192 |
| rs2230912 | 1,405 | 0.217 | – | – | 0.017 |
| rs3751143 | 1,513 | 0.136 | 0.205 | 0.239 | 0.067 |
| rs1653624 | 1,729 | 0.017 | – | – | – |
CEU CEPH Utah Residents with Northern and Western European Ancestry, CHB Han Chinese in Beijing, China, JPT Japanese in Tokyo, Japan, YRI, Yoruba in Ibadan, Nigeria
P2X7R and human disease
Genetic association studies of human disease with a complex pattern of inheritance have yielded valuable insights into the involvement of unsuspected genes. Several genetic association case–control studies using synonymous and non-synonymous SNPs have revealed a role for P2RX7 as a susceptibility gene in mood disorders and in susceptibility to infections with intracellular pathogens such as tuberculosis. Three large studies in a total of 2,500 patients with either bipolar disorder or major depressive disorder [16–18] have found a significant disease association of a non-synonymous SNP (rs2230912) within the coding region of the P2RX7 gene. This SNP codes for Gln460 to Arg in the carboxyl terminus of the receptor protein, but its functional effect on receptor function is not fully defined. P2X7R has also been shown to be important in the killing of phagocytosed Mycobacterium tuberculosis by extracellular ATP. Extensive in vitro data have shown that mycobacteria can survive following uptake by macrophages, but that a bactericidal pathway activated by extracellular ATP via the P2X7 receptor leads both to death of the mycobacteria and subsequent apoptosis of the infected macrophage [19–23]. Epidemiological studies have confirmed the role of P2X7R in the control of tuberculosis. Thus, inheritance of the 1513A>C loss-of-function polymorphic variant of P2RX7 confers a 3.5-fold increased lifetime risk of reactivating latent (primary) tuberculosis in two cohorts of south-east Asian migrants to Australia [22], while a Mexican study has confirmed 1513A>C as a susceptibility factor for reactivating tuberculosis [24]. A role for P2X7R has also been shown in resistance to infection with Chlamydia trachomatis [25, 26] although there have been no large association studies to date studying Chlamydia infection and SNPs in P2RX7.
P2X7R may also play a role in bone disease and recent studies have suggested its function as a mechano-transducer in osteocytes [27] and thus inheritance of the loss-of-function P2X7R variants Glu496 to Ala and Ile568 to Asn is associated with increased fracture risk in post-menopausal females [28].
Defining haplotype structure in P2RX7 using pairwise linkage disequilibrium
Defining linkage disequilibrium (LD) patterns in P2RX7 for use in association studies is essential for selecting a minimum set of SNPs for genotyping and to show the extent of a region to be examined for functional variants. These data are also helpful to explain failure of previous association studies to detect the involvement of P2RX7 if the chosen marker SNPs are not in LD with functional variants. We investigated pairwise LD using ten non-synonymous SNPs to define haplotype blocks in an Australian Caucasian population sample. The program Haploview (http://www.broad.mit.edu/mpg/haploview/index.php) was used to test for conformance with Hardy–Weinberg equilibrium, calculate D′ and r2 measures of LD between SNP pairs and to identify haplotype blocks of strong LD. In addition, we compared these data with patterns of variation for P2RX7 found in the International HapMap Project (http://www.hapmap.org) database.
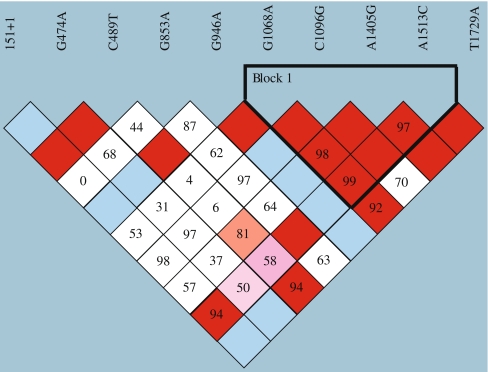
Distributions of genotype frequencies of the ten SNPs in the Australian Caucasians were all in Hardy–Weinberg equilibrium, and minor allele frequencies are summarized in Table 1. A haplotype block was found to include base positions 1068, 1096, 1405, and 1513 (Fig. 1) and this block overlaps that previously published [16, 17]. It is likely that the base position 1729 lies within this block, but the low frequency of 1729A minor allele precluded its inclusion. Analysis of the HapMap International Project (http://www.hapmap.org) data confirmed significant pairwise LD between base positions 1068 and 1096, 1405, and 1513 (not shown). For these four SNPs 99.9% of our Australian Caucasian subjects had one of five haplotypes: GCAA, ACAA, GCAC, ACGA, and GGAA (Table 3). The 489 base position was not part of this haplotype block but was in high LD with 1405 (D′ = 0.81) and 1729 (D′ = 0.94). LD between 489 and 1405 was confirmed in the CEU population using the HapMap (http://www.hapmap.org) data. Analysis of 1,166 subjects that were genotyped at both 489 and 1405 and 1,170 subjects genotyped at both 489 and 1513 confirmed that 1405G and 1513C minor alleles were associated with the 489T minor allele (Table 4).
Fig. 1.
HaploView analysis of pairwise linkage disequilibrium in the P2RX7 gene using ten marker SNPs that change receptor function. The colours represent the relative D’/LOD score where bright red is D’=1; LOD≥2 and blue D’=1; LOD<2, shades of pink D’<1; LOD≥2, white D’ < 1; LOD < 2. Numbers represent D’ scores for pair-wise linkage disequilibrium
Table 3.
Five haplotypes within the P2RX7 genomic block which make up 99.9% of Australian Caucasian subjects
| 1068 | 1096 | 1405 | 1513 | Frequency |
|---|---|---|---|---|
| G | C | A | A | 0.341 |
| A | C | A | A | 0.226 |
| G | C | A | C | 0.185 |
| A | C | G | A | 0.173 |
| G | G | A | A | 0.075 |
Table 4.
Genotype of Australian Caucasian subjects who have been typed at (a) both 489 and 1,405 bp of the gene (n = 1,166) and (b) both 489 and 1,513 bp of the gene (n = 1,170)
| 489 | ||||
|---|---|---|---|---|
| CC | CT | TT | ||
| 1,405 | AA | 305 | 358 | 100 |
| AG | 21 | 240 | 108 | |
| GG | 0 | 4 | 30 | |
| 1,513 | AA | 280 | 392 | 107 |
| AC | 46 | 192 | 102 | |
| CC | 0 | 22 | 29 | |
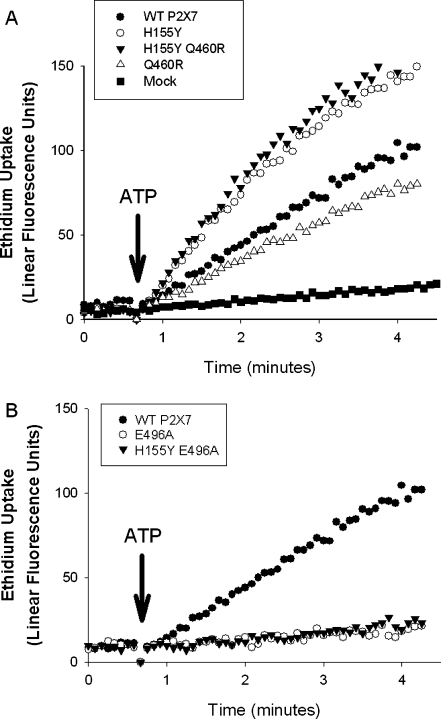
We have investigated these particular strong pairwise LD relationships by transfecting HEK293 cells with constructs carrying a combination of His155 to Tyr and Gln460 to Arg mutations and a combination of His155 to Tyr and Glu496 to Ala mutations. This approach allows an assessment of whether a gain-of-function polymorphism can modulate the functional effect of a second co-inherited polymorphism. Figure 2 shows that the gain-of-function Tyr155 mutation increases ATP-induced ethidium uptake by approximately 50% over a wild-type receptor, while the Arg460 mutation shows reduced ethidium uptake. The combination shows that the Tyr155 rescued the function of Arg460 (Fig. 2a), whereas the Tyr155 mutation could not rescue the loss-of-function seen with the Ala496 mutation (Fig. 2b).
Fig. 2.
ATP-induced ethidium uptake into HEK293 cells transfected with P2X7 constructs mutated at polymorphic positions that alter function. a Gain-of-function mutation His155 to Tyr is shown either on a wild-type background or combined with the Gln460 to Arg (Q460R) variation (H155Y Q460R). b Loss-of-function mutation Glu496 to Ala is shown either on a wild-type background (E496A) or combined with the His155 to Tyr (H155Y) variation (H155Y E496A)
In summary, strong LD exists between four non-synonymous P2RX7 SNPs that form five haplotypes in 99.9% of Caucasian subjects. Significantly, SNPs within this block are in high LD with the 489 base position (His155 to Tyr) functional variant. Analysis of 1405A>G and 1513A>C revealed both minor alleles to have originally occurred on a 489T background allele. Individual P2RX7 SNPs should not be considered in isolation and interaction of combinations of functional variants within P2X7R must be considered when analyzing P2X7R studies. These data are important for use in interpreting results of association studies, as SNPs that alter P2X7R channel and pore function are candidates for disease-associated variants. In addition, genotyping costs of future association studies using Caucasian subjects can be reduced by using tag SNPs that identify each of the five haplotypes with LD between four SNPs.
References
- 1.International Human Genome Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431(7011):931–45 [DOI] [PubMed]
- 2.The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437(7063):1299–1320 [DOI] [PMC free article] [PubMed]
- 3.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P et al (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 62(3):676–689 [DOI] [PMC free article] [PubMed]
- 4.Burn J, Chapman P, Delhanty J, Wood C, Lalloo F, Cachon-Gonzalez MB et al (1991) The UK Northern region genetic register for familial adenomatous polyposis coli: use of age of onset, congenital hypertrophy of the retinal pigment epithelium, and DNA markers in risk calculations. J Med Genet 28(5):289–296 [DOI] [PMC free article] [PubMed]
- 5.The Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447(7145):661–678 [DOI] [PMC free article] [PubMed]
- 6.Buell GN, Talabot F, Gos A, Lorenz J, Lai E, Morris MA et al (1998) Gene structure and chromosomal localization of the human P2X7 receptor. Receptors Channels 5(6):347–354 [PubMed]
- 7.Gu BJ, Zhang W, Worthington RA, Sluyter R, Dao-Ung P, Petrou S et al (2001) A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J Biol Chem 276(14):11135–11142 [DOI] [PubMed]
- 8.Boldt W, Klapperstuck M, Buttner C, Sadtler S, Schmalzing G, Markwardt F (2003) Glu496Ala polymorphism of human P2X7 receptor does not affect its electrophysiological phenotype. Am J Physiol Cell Physiol 284(3):C749–C756 [DOI] [PubMed]
- 9.Wiley JS, Dao-Ung LP, Li C, Shemon AN, Gu BJ, Smart ML et al (2003) An Ile-568 to Asn polymorphism prevents normal trafficking and function of the human P2X7 receptor. J Biol Chem 278(19):17108–17113 [DOI] [PubMed]
- 10.Smart ML, Gu B, Panchal RG, Wiley J, Cromer B, Williams DA et al (2003) P2X7 receptor cell surface expression and cytolytic pore formation are regulated by a distal C-terminal region. J Biol Chem 278(10):8853–8860 [DOI] [PubMed]
- 11.Gu BJ, Sluyter R, Skarratt KK, Shemon AN, Dao-Ung LP, Fuller SJ et al (2004) An Arg307 to Gln polymorphism within the ATP-binding site causes loss of function of the human P2X7 receptor. J Biol Chem 279(30):31287–31295 [DOI] [PubMed]
- 12.Skarratt KK, Fuller SJ, Sluyter R, Dao-Ung LP, Gu BJ, Wiley JS (2005) A 5′ intronic splice site polymorphism leads to a null allele of the P2X7 gene in 1–2% of the Caucasian population. FEBS Lett 579(12):2675–2678 [DOI] [PubMed]
- 13.Shemon AN, Sluyter R, Fernando SL, Clarke AL, Dao-Ung LP, Skarratt KK et al (2006) A Thr357 to Ser polymorphism in homozygous and compound heterozygous subjects causes absent or reduced P2X7 function and impairs ATP-induced mycobacterial killing by macrophages. J Biol Chem 281(4):2079–2086 [DOI] [PubMed]
- 14.Cabrini G, Falzoni S, Forchap SL, Pellegatti P, Balboni A, Agostini P et al (2005) A His-155 to Tyr polymorphism confers gain-of-function to the human P2X7 receptor of human leukemic lymphocytes. J Immunol 175(1):82–89 [DOI] [PubMed]
- 15.Denlinger LC, Coursin DB, Schell K, Angelini G, Green DN, Guadarrama AG et al (2006) Human P2X7 pore function predicts allele linkage disequilibrium. Clin Chem 52(6):995–1004 [DOI] [PubMed]
- 16.Barden N, Harvey M, Gagne B, Shink E, Tremblay M, Raymond C et al (2006) Analysis of single nucleotide polymorphisms in genes in the chromosome 12Q24.31 region points to P2RX7 as a susceptibility gene to bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet 141B(4):374–382 [DOI] [PubMed]
- 17.Lucae S, Salyakina D, Barden N, Harvey M, Gagne B, Labbe M et al (2006) P2RX7, a gene coding for a purinergic ligand-gated ion channel, is associated with major depressive disorder. Hum Mol Genet 15(16):2438–2445 [DOI] [PubMed]
- 18.McQuillin A, Bass NJ, Choudhury K, Puri V, Kosmin M, Lawrence J, et al (2008) Case–ontrol studies show that a non-conservative amino-acid change from a glutamine to arginine in the P2RX7 purinergic receptor protein is associated with both bipolar- and unipolar-affective disorders. Mol Psychiatry. doi:1038/mp.2008.6 [DOI] [PubMed]
- 19.Saunders BM, Fernando SL, Sluyter R, Britton WJ, Wiley JS (2003) A loss-of-function polymorphism in the human P2X7 receptor abolishes ATP-mediated killing of mycobacteria. J Immunol 171(10):5442–5446 [DOI] [PubMed]
- 20.Fairbairn IP, Stober CB, Kumararatne DS, Lammas DA (2001) ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X(7)-dependent process inducing bacterial death by phagosome–lysosome fusion. J Immunol 167(6):3300–3307 [DOI] [PubMed]
- 21.Kusner DJ, Adams J (2000) ATP-induced killing of virulent Mycobacterium tuberculosis within human macrophages requires phospholipase D. J Immunol 164(1):379–388 [DOI] [PubMed]
- 22.Fernando SL, Saunders BM, Sluyter R, Skarratt KK, Goldberg H, Marks GB et al (2007) A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am J Respir Crit Care Med 175(4):360–366 [DOI] [PubMed]
- 23.Lammas DA, Stober C, Harvey CJ, Kendrick N, Panchalingam S, Kumararatne DS (1997) ATP-induced killing of mycobacteria by human macrophages is mediated by purinergic P2Z(P2X7) receptors. Immunity 7(3):433–444 [DOI] [PubMed]
- 24.Nino-Moreno P, Portales-Perez D, Hernandez-Castro B, Portales-Cervantes L, Flores-Meraz V, Baranda L et al (2007) P2X7 and NRAMP1/SLC11 A1 gene polymorphisms in Mexican mestizo patients with pulmonary tuberculosis. Clin Exp Immunol 148(3):469–477 [DOI] [PMC free article] [PubMed]
- 25.Coutinho-Silva R, Stahl L, Raymond MN, Jungas T, Verbeke P, Burnstock G et al (2003) Inhibition of chlamydial infectious activity due to P2X7R-dependent phospholipase D activation. Immunity 19(3):403–412 [DOI] [PubMed]
- 26.Coutinho-Silva R, Perfettini JL, Persechini PM, Dautry-Varsat A, Ojcius DM (2001) Modulation of P2Z/P2X(7) receptor activity in macrophages infected with Chlamydia psittaci. Am J Physiol Cell Physiol 280(1):C81–C89 [DOI] [PubMed]
- 27.Panupinthu N, Rogers JT, Zhao L, Solano-Flores LP, Possmayer F, Sims SM et al (2008) P2X7 receptors on osteoblasts couple to production of lysophosphatidic acid: a signaling axis promoting osteogenesis. J Cell Biol 181(5):859–871 [DOI] [PMC free article] [PubMed]
- 28.Ohlendorff SD, Tofteng CL, Jensen JE, Petersen S, Civitelli R, Fenger M et al (2007) Single nucleotide polymorphisms in the P2X7 gene are associated to fracture risk and to effect of estrogen treatment. Pharmacogenet Genomics 17(7):555–567 [DOI] [PubMed]