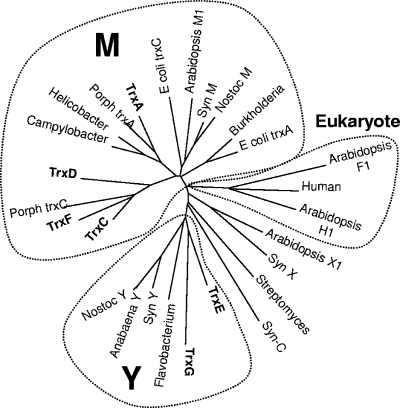
FIG. 1.
Phylogenetic comparison of 27 Trx proteins from diverse sources. ClustalW was used to align protein sequences for 27 Trx proteins. The unrooted bootstrap consensus tree was inferred using the minimum-evolution method with 500 bootstrap replicates (41). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Phylogenetic analyses were conducted in MEGA4 (50). The following sequences with accession numbers were used: Burkholderia, YP_333769; human, NP_003320; E. coli TrxA, AAA67582; Porph TrxC, P. gingivalis AAQ65495; Syn M, Synechococcus ZP_01124485; Anabaena Y, ABA23368; Flavobacterium, CAL43878; Nostoc Y, NP_485933; Syn Y, Synechocystis NP_442168; Campylobacter, YP_178167; Helicobacter, NP_223481; Porph TrxA, P. gingivalis NP_904389; Streptomyces, CAB72414; TrxA, YP_210347; TrxC, YP_212311; TrxD, YP_211860; TrxE, YP_212629; TrxF, YP_212630; TrxG, YP_210941; Arabidopsis H1, CAA78462; Arabidopsis F1, AAD35003; Arabidopsis X1, NP_564566; Syn X, Synechocystis NP_440611; Syn C, Synechocystis NP_439965; E. coli TrxC, NP_417077; Arabidopsis M1, AAF15948; Nostoc M, NP_485906.