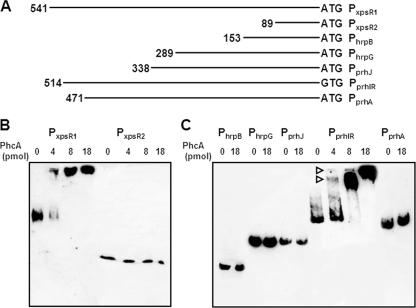
FIG. 3.
Gel mobility shift assay for PhcA. (A) Biotinylated promoter DNA fragments are as follows: PxpsR1, xpsR containing the PhcA binding region; PxpsR2, xpsR lacking this region; PhrpB, hrpB; PhrpG, hrpG; PprhJ, prhJ; PprhIR, prhIR; PprhA, prhA. The length of the fragment (bp) is shown at the left end of each line. Next to the 3′ end is a start codon marked ATG or GTG. (B) Biotinylated xpsR promoter fragments were mixed with purified PhcA with C-terminal His tag (0 to 18 pmol), separated on a polyacrylamide gel, and transferred onto a positively charged nylon membrane. The membrane was treated with streptavidin-horseradish peroxidase conjugate, and labeled DNA fragments were visualized with Chemi-Lumi One (Nakarai Tesque, Kyoto, Japan). One microgram of poly(dI·dC) was added as nonspecific competitor DNA. (C) Biotinylated promoter fragments of genes belonging to the hrp gene regulatory cascade were incubated with His-tagged PhcA. Promoters PhrpB, PhrpG, PprhJ, PprhIR, and PprhA are shown on the top. Open triangles indicate the DNA fragments forming complexes with PhcA.