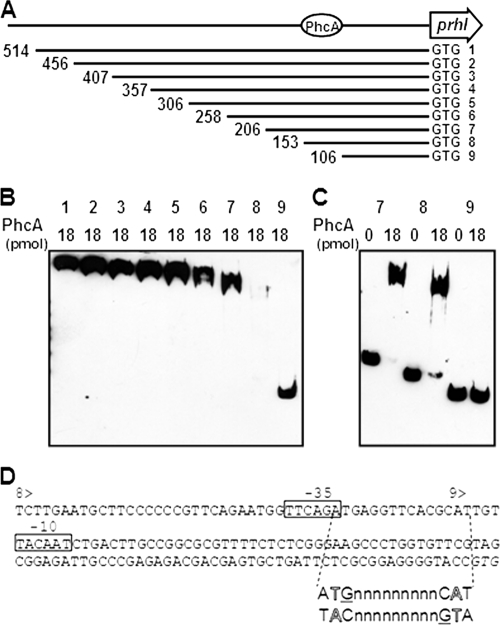
FIG. 4.
Gel mobility shift assay for PhcA binding to the prhIR promoter region. (A) Schematic drawing of prhIR promoter deletion. 1, PprhIR; 2, PprhIR2; 3, PprhIR3; 4, PprhIR4; 5, PprhIR5; 6, PprhIR6; 7, PprhIR7; 8, PprhIR8; 9, PprhIR9. The length of fragment (bp) is shown at the left end of each line. Next to the 3′ end is a start codon marked GTG. The putative PhcA binding site is indicated by a circle. (B) Biotinylated prhIR fragments with several deletions were mixed with 18 pmol of purified His-tagged PhcA and separated by SDS-PAGE. DNA bands were visualized as described for Fig. 3. Numbers above the lanes indicate the promoter fragments represented in panel A. All DNA fragments were incubated with 18 pmol of PhcA. (C) Biotinylated prhIR fragments PprhIR7, PprhIR8, and PprhIR9 were mixed with either 0 or 18 pmol of purified His-tagged PhcA. Numbers above the lanes indicate the promoter fragments represented in panel A. (D) Nucleotide sequence of the prhIR promoter region. 8> and 9> indicate the beginnings of PprhIR8 and PprhIR9, respectively. Putative −35 and −10 sequences of σ70 are boxed. A hypothetical recognition site sequence is shown below to illustrate the interrupted dyadic sequence and T-N11-A motif (outlined letters); guanines expected to be directly involved in binding are underlined. The initiation codon is shown in italics.