Abstract
Several drug development strategies, including optimization of new antimalarial drug combinations, have been used to counter malaria drug resistance. We evaluated the malaria Sybr green I-based fluorescence (MSF) assay for its use in in vitro drug combination sensitivity assays. Drug combinations of previously published synergistic (atovaquone and proguanil), indifferent (chloroquine and azithromycin), and antagonistic (chloroquine and atovaquone) antimalarial drug interactions were tested against Plasmodium falciparum strains D6 and W2 using the MSF assay. Fifty percent inhibitory concentrations (IC50s) were calculated for individual drugs and in fixed ratio combinations relative to their individual IC50s. Subsequent isobologram analysis and fractional inhibitory concentration determinations demonstrated the expected drug interaction pattern for each combination tested. Furthermore, we explored the ability of the MSF assay to examine mixed parasite population dynamics, which are commonly seen in malaria patient isolates. Specifically, the capacity of the MSF assay to discern between single and mixed parasite populations was determined. To simulate mixed infections in vitro, fixed ratios of D6 and W2 strains were cocultured with antimalarial drugs and IC50s were determined using the MSF assay. Dichotomous concentration curves indicated that the sensitive and resistant parasites composing the genetically heterogeneous population were detectable. Biphasic analysis was performed to obtain subpopulation IC50s for comparison to those obtained for the individual malaria strains alone. In conclusion, the MSF assay allows for reliable antimalarial drug combination screening and provides an important method to discern between homogenous and heterogeneous parasite populations.
Malaria is a severe global health problem that is compounded by the emergence of drug-resistant parasites. The emergence of these multidrug-resistant Plasmodium species, particularly P. falciparum, has made decisions regarding malaria chemoprophylaxis and treatment more complicated. Furthermore, it is predicted that increased incidences of clinical infections and subsequent deaths are likely as the rapid spread of resistant parasites occurs (15, 23, 35, 39). Several drug development strategies have been used to counter malaria drug resistance, including optimization of new antimalarial drug combinations. Several groups have examined drug combinations in laboratory settings that may have promising efficacies in clinical settings (5, 7, 30). Artemisinin-based combination therapies, one of the most successful therapeutic combinations, are currently used in areas where malaria is endemic (39). However, it is predicted that even with an aggressive prophylactic and treatment campaign, resistance to these drugs will certainly emerge (8). In fact, resistance to these combinations has already been observed. Wongsrichanalai and others reported the decreasing efficacy of the artesunate-mefloquine combination on the Cambodian-Thai border (41). Thus, there is a need to discover novel combinations between existing antimalarials and/or new chemical entities that can be used in the treatment of severe malaria.
In addition to resistance to antimalarial combination therapies, there is also a concern with Plasmodium mixed infections. A mixed infection is defined as an infection with more than one type of species or genotype of Plasmodium (24). Although highly understudied, the implications of a mixed infection are profound. Mixed infections can cause a relapse as a result of emergence of the resistant subpopulation of parasites after the sensitive subpopulation has been eradicated by drug therapy. The existence of a resistant population may be a result of both divergent evolution, where parasites have acquired resistance mechanisms, and/or two cohabitating parasites when the individual is infected (16, 24). This phenomenon has been observed in areas of malaria endemicity in Africa and Southeast Asia, where the mixed-infection prevalence is as high as 30% (24). However, there has been conflicting evidence as to the true frequency of Plasmodium mixed infections (27, 34). Furthermore, this problem is confounded by the inability to properly identify and differentiate Plasmodium mixed infections.
The [3H]hypoxanthine incorporation assay has been used as the gold standard in P. falciparum drug susceptibility testing (11). Despite the assay's reliability and accuracy, it is very expensive, involves multiple processing steps, and requires special handling and waste disposal procedures. In contrast, dye-based technologies, such as those using 4′,6-diaminino-2-phenylindole, Pico green, YOYO-1, and Sybr green, have been shown to have comparable results to radioactive assays (2, 10, 17, 19, 36, 38). Many of these assays use DNA dye intercalation, which accurately measures parasite growth. Use of these assays has increased because they are relatively simple and inexpensive to run compared to their radioactive and enzyme-linked immunosorbent assay-based counterparts.
While several in vitro drug sensitivity assays have been used to analyze antimalarial drug interactions, the ability of the malaria Sybr green I-based fluorescence (MSF) assay for this purpose has not been fully characterized. The [3H]hypoxanthine incorporation assay has been shown to detect differences in susceptibility patterns as well as being able to identify drug interactions (11, 30). However, radioactivity usage makes it costly and difficult to routinely use in research and clinical settings, particularly in a resource-limited environment. The MSF assay utilizes the Sybr green I dye (Invitrogen, San Diego, CA), which is relatively inexpensive and has been shown to reliably measure P. falciparum in vitro drug sensitivities. As the prevalence of Plasmodium mixed infections increases, there is a need for an assay that can reliably identify and differentiate drug-sensitive subpopulations in a particular infection. In this study, we examined the capability of the MSF assay to determine drug interactions and discern between single and mixed P. falciparum populations.
MATERIALS AND METHODS
Malaria Sybr green I-based fluorescence assay setup.
The MSF assay was performed as described by Smilkstein et al. (36) and as modified by Johnson et al. (17). Briefly, D6 (CDC/Sierra Leone) and W2 (CDC/Indochina) P. falciparum strains were maintained in continuous long-term cultures in tissue culture medium, as previously described (11, 26). Cultures and assays were conducted at 37°C under a humidified atmosphere of 5% CO2 and 5% O2, with a balance of N2 using either tissue culture medium or folic acid-free medium. Parasites at 1% parasitemia and 2% hematocrit were added to predosed 96-well plates and incubated for 72 h. Lysis buffer (20 mM Tris-HCl, 5 mM EDTA, 0.008% saponin [Sigma-Aldrich], and 0.08% Triton X [Sigma-Aldrich]) with Sybr green I dye (Invitrogen) was subsequently added to the plates and incubated for 1 h at ambient room temperature. Fluorescence was read using the GENios Plus plate reader (Tecan, Research Triangle Park, NC). The plates were examined for relative fluorescence units (RFUs) per well using the Tecan Genios Plus reader. The drug concentrations (x values) were transformed using the formula X = log[X] and plotted against the RFUs (y values). The data were then analyzed with Prism 4.0 (GraphPad Software, Inc., San Diego, CA) by nonlinear regression (sigmoidal dose-response/variable slope equation) to yield drug 50% inhibitory concentrations (IC50s).
MSF drug combination assay.
Fixed ratio combinations of various antimalarial drugs were tested against P. falciparum D6 and W2 strains as previously described by Orht et al. (30). Briefly, each drug was tested alone and at fixed ratios with 12 twofold serial dilutions and evaluated using the standard MSF assay conditions described above. The IC50s were calculated for each drug alone and for their respective fixed concentration ratios. The individual and sum 50% fractional inhibitory concentrations (FIC50 and ∑FIC50, respectively) were determined as previously described (6). Isobolograms were constructed from the FIC50s of drug A and drug B at the tested fixed concentration ratios, for which a straight line represents indifference (∑FIC = 1), a concave line denotes a trend toward synergy (∑FIC < 1) or synergy (∑FIC ≤ 0.5), and a convex curve represents a trend toward antagonism (∑FIC > 1) or antagonism (∑FIC ≥ 2 to 4). The IC50s of each drug in the test combination were standardized by allocating the value of 1 to each drug that was tested alone and prorated values for each fixed concentration ratio.
MSF mixed-infection assay.
Mixed parasite infections were simulated in vitro by coculturing D6 and W2 strains in various fixed ratios to create heterogeneous parasite populations in a 96-well format. Dichotomous response curves were plotted by standardizing RFU values to relative growth percentages. For drugs that displayed a dichotomous response, subpopulation IC50s were determined using a modification of the logistic-logarithmic function previously described by Oduola et al. (29). Briefly, biphasic curves were generated for each fixed ratio populations, with each phase of the curve analyzed as a separate IC50 curve. These individual IC50 curves and combined IC50s were determined using GraphPad Prism. The concentration-response profile of each parasite strain alone was then compared to responses of the coculture populations and its respective subpopulation components. As a negative control, artemisinin monophasic curves were generated at fixed parasite ratios as previously described.
Statistical analyses.
Data were analyzed by analysis of variance and Tukey's multiple comparison test (Sigma Stat 3.1; SPSS, Inc., Chicago, IL).
RESULTS
MSF combination testing assessment.
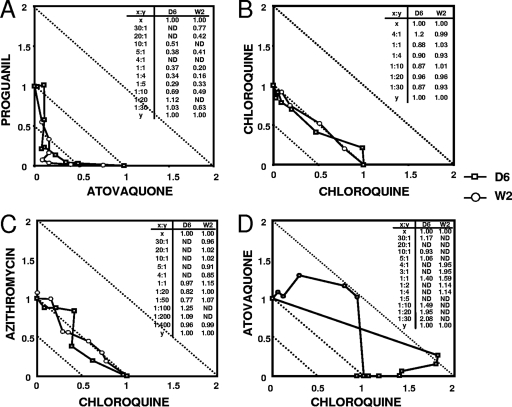
To assess the MSF assay's ability to determine synergistic, indifferent, and antagonistic in vitro antimalarial drug interactions we tested fixed ratio combinations of atovaquone-proguanil, chloroquine-azithromycin, and chloroquine-atovaquone, respectively, against P. falciparum D6 and W2 strains. Analysis of drug interactions was performed by the method described by Berenbaum (6). Proguanil and atovaquone revealed a synergistic interaction and chloroquine and azithromycin showed indifference, while chloroquine and atovaquone had antagonistic interactions in both D6 and W2 strains (Fig. 1A, C, and D). The ΣFIC50s of tested fixed drug ratio combinations are presented within each figure. The combination of chloroquine with itself served as an experimental drug combination control, displaying indifference (Fig. 1B).
FIG. 1.
Isobologram analysis of IC50s of single and combination drugs at fixed concentrations. ΣFIC50 values at fixed drug concentrations are shown in inset tables. Graphs display the following interactions: synergism (A), indifference (B and C), antagonism (D). ND, not determined. Data are from a representative of three to five experiments.
Mixed-infection study.
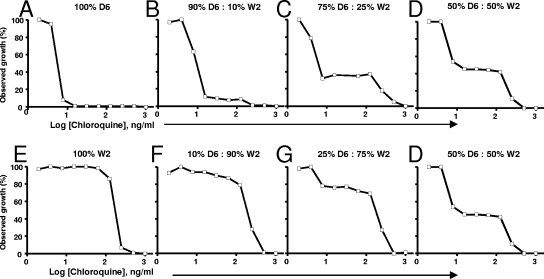
To assess the MSF's capability to discern between single and mixed populations, we tested various fixed ratios of D6 and W2 against various antimalarial drugs. The mixed strain populations were allowed to grow for 24, 48, 72, and 96 h to observe whether overgrowth of one strain occurred. For the drugs that displayed a dichotomous response, we derived the same biphasic curve at times of ≥72 h (data not shown). As seen in Fig. 2, dichotomous concentration curves indicated that the chloroquine-sensitive and -resistant parasite subpopulations composing a genetically heterogeneous population grown at 72 h were detectable using the MSF assay. Figure 2 shows alterations of the dichotomous curve depending on the susceptibility profile of the populations. D6 is sensitive to chloroquine (IC50, 4.66 ng/ml), and increasing ratios of W2 (Fig. 2D, G, F, and E) shift the dichotomous response curve toward a more resistant profile (Fig. 2). The reverse response was also observed when increasing ratios of D6 were present in the population. A plateauing effect was also observed with D6 and W2 mixed population curves, in comparison to their corresponding single-population monophasic curves. Furthermore, Fig. 2 shows that for insets B, C, and D, where the ratio of D6 to W2 was 90:10, 75:25, and 50:50, the corresponding D6 subpopulation growth percentages were approximately 90%, 75%, and 50%. Subsequently, the remaining subpopulation corresponds to W2. The level where the plateau occurs (Fig. 2B to D, F, and G) allows for the estimation of subpopulation percentages within the population. Furthermore, this plateau population corresponds to the more resistant population, since the growth is restricted only at high chloroquine concentrations. Finally, the biphasic curves also serve as an internal control, confirming that our data are comparable to those published by Oduola et al. (29). In addition, our assay system confirms that D6 and W2 have similar growth rates in our assay system, as shown in Fig. 2, where the observed percent growth is correlated to subpopulation parasite levels as previously reported (12, 37).
FIG. 2.
In vitro susceptibilities of mixed population cultures against chloroquine. IC50 curves are shown for 100% D6 (A) and 100% W2 (E), 90% D6 and 10% W2 (B), 10% D6 and 90% W2 (F), 75% D6 and 25% W2 (C), 25% D6 and 75% W2 (G), 50% D6 and 50% W2 (D). Data are from a representative of four to six experiments.
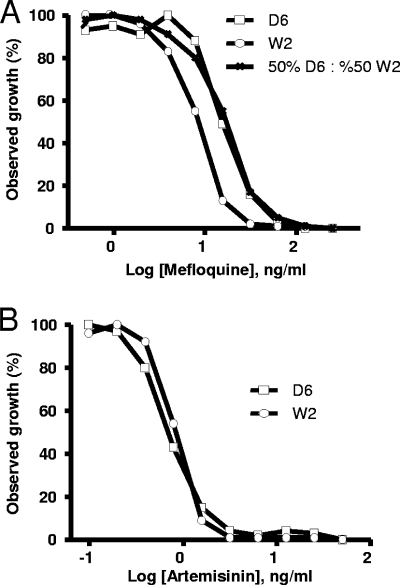
Chloroquine IC50 determinations for each respective population and subpopulation are presented in Table 1. Total population IC50s reflect the biphasic curve obtained, displaying a statistically significant increase in IC50 observed at the 25:75 D6 to W2 mixture compared to that with 100% D6. Subpopulation IC50s for D6 and W2 were not statistically different from those obtained from individual IC50s of D6 or W2 alone (Table 1). When the parasite composition was 90:10 or vice versa, an IC50 for the smaller subpopulation could not be derived because the points obtained did not converge into a discernible dose-response curve for IC50 determinations. We also tested these fixed ratios of D6 and W2 against mefloquine, a drug to which the two tested parasite strains display a narrower difference in IC50s than chloroquine (9.4 and 248.5 ng/ml for chloroquine versus 43.7 and 16.2 ng/ml for mefloquine) (17). Figure 3A shows the drug response to mefloquine by a 50:50 D6:W2 population as well as drug responses by the individual strains. D6 was resistant to mefloquine (IC50, 43.7 ng/ml) and any W2 population containing as little as 10% D6 was enough to shift the curve to a more resistant profile (data not shown). Consequently, at a 50:50 D6:W2 ratio, the generated curve mirrored the one derived from D6 alone, resulting in a monophasic curve. Taken together, dichotomous curves could not be derived from the mixed population of D6 and W2 treated with mefloquine.
TABLE 1.
Population and subpopulation chloroquine IC50 results
| D6:W2 ratio | Chloroquine IC50 (ng/ml) fora:
|
||
|---|---|---|---|
| Total population | D6 subpopulation | W2 subpopulation | |
| 100:0 | 9.1 | ||
| 90:10 | 8.3 | 5.9 | DNCb |
| 75:25 | 10.0 | 3.3 | 211.5 |
| 50:50 | 30.0 | 6.4 | 244.4 |
| 25:75 | 80.4* | 5.9 | 278.5 |
| 10:90 | 198.1* | DNCb | 220.1 |
| 0:100 | 210.3* | ||
Data are averages from four to six experiments. *, value is statistically significantly different (P < 0.05) from D6:W2 (100:0).
DNC, did not converge (for the subpopulation IC50 determination).
FIG. 3.
In vitro susceptibilities of mixed population cultures against mefloquine (A) and artemisinin (B). Data are from a representative of four to six experiments.
Finally, as a control, we also performed the same mixed parasite ratio experiment with artemisinin, to which D6 and W2 have similar IC50s (1.0 ng/ml). Figure 3B shows monophasic curves to artemisinin obtained with the D6 and W2 strains individually. As expected, dichotomous curves could not be generated for artemisinin (data not shown), as the monophasic curves obtained for this drug were identical. These data demonstrate that a dichotomous response cannot be derived if the susceptibility profiles of the subpopulations are similar.
DISCUSSION
Previously, we compared the Sybr green I-based fluorescence (MSF) assay to the [3H]hypoxanthine assay and showed that the MSF assay has the sensitivity and reproducibility required for malaria drug susceptibility testing (17). This has led us to further assess the MSF assay as a rapid method for screening potential new drug combinations. In this report, we specifically examined the MSF assay's ability to detect previously characterized in vitro drug interactions. Fixed ratio combinations of compounds with known antimalarial activities were tested against P. falciparum D6 and W2 strains. Our data confirmed findings by others who used the [3H]hypoxanthine assay, specifically, Canfield et al., who described a synergistic interaction between proguanil and atovaquone and antagonism between chloroquine and atovaquone (7). Also using the [3H]hypoxanthine assay, Nakornchai and konthiang found an indifferent interaction between chloroquine and azithromycin (28). Overall, these data show that the MSF assay can be a useful tool to assess the efficacy of combining drugs with known activities with each other, as well as determining the efficacies of existing therapeutics with novel drug entities in a high-throughput screening format. In fact, we also performed this type of combination testing on 384-well plates and observed results comparable to those in the 96-well plates (data not shown). Performing this assay in a 384-well format will allow multiple drugs to be tested simultaneously and facilitate the testing of a broader range of drug concentrations. As numerous laboratories continue to screen drugs against parasites for either surveillance of drug resistance or drug discovery efforts, there is a dire need to standardize assays to evaluate the efficacies of drug combinations (5). The ease of use and comparability with the [3H]hypoxanthine assay make the MSF assay a viable format for standardized evaluation of in vitro antimalarial activity.
Most naturally occurring malaria infections are composed of mixed subpopulations with different drug susceptibilities (22, 27, 32, 42). Thus, there is a need to identify these drug-resistant subpopulations to prevent treatment failures. In our previous work, we showed that the MSF assay can reliably monitor drug-resistant parasites (17). In this study, we further examined the MSF assay's ability to discern between single and mixed P. falciparum parasite populations, using the method adapted from Oduola et al. to create dichotomous curves from a variety of drugs, and total population and individual subpopulation IC50s were calculated (29). We found that our data were in agreement with the findings of Oduola et al., as the curve they obtained from a 50:50 mixture of D6 and W2 reflected the one presented in Fig. 2D here, which indicates that the MSF assay results are similar to those of the [3H]hypoxanthine assay (29). In addition to the correlation between the dose-response curve plateau and the subpopulation percentages, our data also indicate that the parasite growth rates are similar and that one subpopulation did not outgrow the other within the time periods assayed. It also suggests that obtaining a biphasic curve confirms the existence of parasite subpopulations in a sample, provided they have different susceptibilities. However, we found that the smaller the difference in sensitivities, the less accurate the MSF assay was in identifying a subpopulation, as evidenced by the monophasic curves obtained with mefloquine. The mixed population monophasic mefloquine curve reflects the one derived from the more resistant strain (D6). Thus, the smaller the difference in IC50s, the more difficult it is to discern between subpopulations by testing with just one drug. This presents a weakness with the MSF assay, for which, in order to determine the presence of mixed populations, testing with multiple drugs is required to ensure coverage of the various drug susceptibilities that exist in nature. With other infectious diseases, such as ones caused by bacteria, the use of drug batteries (antibiograms) often allows for better characterization of the susceptibility profile of the organism (9). Particularly with bacterial anaerobic infections, subpopulations with varied antibacterial susceptibilities are observed. Although empirical treatment for these types of infections is prescribed due to the length of time it takes to culture these organisms in determining their drug susceptibility, the failure rate is high due to the proliferation of resistance mechanisms (18). CLSI recommends drug battery testing to allow for informed decisions regarding treatment (9). The same rationale may apply in P. falciparum mixed infections.
Nevertheless, the biphasic curve analysis can predict with reasonable accuracy the relative compositions of parasite subpopulations within a given sample. The occurrence of mixed infections has profound implications in clinical testing (21, 40, 42). This analysis can also be tailored to populations that have more than two types of strains, resulting in triphasic or even quadriphasic curves.
Liu et al. also conducted a similar study where they used DD2 and HB3 to determine the effects of mixed infections in drug testing and genotyping (20). It is interesting that they found a shift in their chloroquine monophasic curve as the resistant parasite percentage increased, while with our assay we observed a dichotomous response to chloroquine. Furthermore, they observed a low percentage of the resistant parasites (as low as 10%) was required to induce a curve shift toward a more resistant profile. This is in contrast to our findings, where we found that this curve shifting was drug specific, as it was monophasic for mefloquine and dichotomous for chloroquine. Although they used different strains in their study, the differences in responses could be accounted for by the genotypic differences inherent in the strains. Even between D6 and W2, there are subtle differences in susceptibility profiles (17) as a result of differing resistance mechanisms inherent in the two strains for particular drugs. Furthermore, our analysis showed an increased percentage of resistant parasites is required before a statistically significant shift in IC50s can be observed with chloroquine. This is in contrast to the findings by Liu et al., where a 10% chloroquine-resistant parasite subpopulation was enough to influence the IC50. However, our findings with the test drug mefloquine are in agreement with Liu et al.'s, as we saw a profound influence of the resistant D6 strain on the overall population IC50, and the shift in the IC50 curves was not gradual but was an all-or-none phenomenon. As mentioned previously, the discrepancies between results for the two studies can be explained by the differences between the strains used.
We also calculated individual subpopulation IC50s from the biphasic curve as described previously and showed that individual subpopulation IC50s can be derived. These subpopulation IC50s serve as controls so that the individual strains can be identified and to rule out effects of coculturing on individual susceptibilities. These values were not statistically different from the IC50s of D6 or W2 alone, indicating that the MSF assay can reliably and accurately calculate subpopulation IC50s. However, our data also showed that when a subpopulation is present in a low proportion (<10%), the MSF assay cannot be used to calculate the IC50 for that subpopulation because the curves do not converge and cannot be used to create an IC50 dose-response curve. This phenomenon is possibly due to the assay's detection limit, where susceptibility of one population ceases to affect the profile of the other subpopulation, effectively masking the existence of that subpopulation. At this low parasite subpopulation level, dominance of a resistance mechanism may not be high enough in the population and may result in the appearance of only one phenotype. The reverse is also possible, where a large susceptible subpopulation may influence the apparent phenotype of the whole population. This finding is similar to that of Liu et al., who reported IC50s with lower subpopulation compositions were not distinguishable from IC50s with the parasite alone (20). Since the conclusion of this work, we have miniaturized our 96-well plate setup to a 384-well plate setup using procedures similar to those of Plouffe et al. (31) and Weisman et al. (38). In performing antimalarial compound susceptibility tests, we have increased our sensitivities and lowered our detection limit fourfold (data not shown) by changing the concentrations of our lysis buffer components. This shows that the MSF assay's capabilities have yet to be determined, and the assay shows great promise in its capabilities for detection and analysis.
The requirement for continuous culture may affect the selection of subpopulations in a clinical sample. Often, clinical strains are culture adapted, resulting in possible loss of the minor subpopulation and consequent misidentification of the population's susceptibility profile (3, 4, 25). However, other studies have also shown that even with culture adaptation, the minor subpopulation is still detectable (25). In fact, the MSF assay has already been used successfully on clinical isolates, and studies have shown that even with patient-to-plate culturing, subpopulations present in proportions as low as 0.1% are still observable with the HRP II enzyme-linked immunosorbent assay and Sybr green I methods (1, 33). Regardless, our data show that our method has quantitative potential. Unless further validations of the method are performed, we currently recommend that the MSF assay be used as only a qualitative assay for mixed infections. This assay needs to be characterized and validated for its ability to quantitatively detect mixed infections within these clinical isolates.
Other groups have noted that a number of malarial infections are heterogeneous in their composition (21, 40). In fact, almost all infections occurring in nature are mixed, requiring a method that can discern subtle changes in resistance patterns. Several investigators have used molecular markers to discern resistance in clinical samples (13, 14). PCR has been shown to be most effective in identifying the presence of multiple strains due to its rapid use and nonculture requirement. The MSF assay, which has been shown to be robust enough for usage in a resource-limited environment, is a significant complement to any drug resistance genotyping method, particularly in identifying the presence of mixed infections. Currently, there are no diagnostic molecular resistance markers for malaria, making recognition of susceptibility profiles difficult with only PCR. Molecular methods, although accurate in detecting the presence of multiple strains, cannot identify or parse relative resistance patterns. By employing the MSF assay, treatment regimens can immediately be adapted upon mixed-infection identification. However, as previously mentioned, the assay requires validation in terms of its quantitative detection and susceptibility testing. The MSF assay has recently been shown to be efficacious in monitoring susceptibilities in clinical samples (33). This capability could be extended to detecting resistant subpopulations in the clinical setting, since the assay has already shown its amenability to field and resource-limited settings.
We have demonstrated that the MSF assay can be used in in vitro drug combination studies and for discerning drug susceptibilities between subpopulations within mixed infections. Not only is the MSF assay successful in identifying drug interactions in drug combination testing in research settings, but also, others in the literature have shown the assay's usefulness in clinical settings (33). In addition, the MSF assay has been tailored to an ultrahigh-throughput format to discover novel chemical entities with antimalarial activity (31). This capability, coupled with findings in this study, should facilitate the discovery of efficacious drug combinations in a much more efficient manner.
Acknowledgments
The opinions or assertions contained herein are our private views and are not to be construed as official or as reflecting views of the Department of the Army or the Department of Defense.
Footnotes
Published ahead of print on 6 April 2009.
REFERENCES
- 1.Bacon, D. J., C. Latour, C. Luca, O. Colina, P. Ringwald, and S. Picot. 2007. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob. Agents Chemother. 51:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baniecki, M. L., D. F. Wirth, and J. Clardy. 2007. High-throughput Plasmodium falciparum growth assay for malaria drug discovery. Antimicrob. Agents Chemother. 51:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basco, L. K., and J. Le Bras. 1994. In vitro susceptibility of Cambodian isolates of Plasmodium falciparum to halofantrine, pyronaridine, and artemisinin derivatives. Ann. Trop. Med. Parasitol. 88:137-144. [DOI] [PubMed] [Google Scholar]
- 4.Basco, L. K., and J. Le Bras. 1990. Reversal of chloroquine resistance with desipramine in isolates of Plasmodium falciparum from Central and West Africa. Trans. R. Soc. Trop. Med. Hyg. 84:479-481. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. 2005. Antimalarial drug synergism and antagonism: mechanistic and clinical significance. FEMS Microbiol. Lett. 253:171-184. [DOI] [PubMed] [Google Scholar]
- 6.Berenbaum, M. C. 1978. A method for testing for synergy with any number of agents. J. Infect. Dis. 137:122-130. [DOI] [PubMed] [Google Scholar]
- 7.Canfield, C. J., M. Pudney, and E. Gutteridge. 1995. Interactions of atovaquone with other antimalarial drugs against Plasmodium falciparum in vitro. Exp. Parasitol. 80:373-381. [DOI] [PubMed] [Google Scholar]
- 8.Chretien, J. P., M. Fukuda, and H. Noedl. 2007. Improving surveillance for antimalarial drug resistance. JAMA 297:2278-2281. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 7th ed., CLSI document M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Corbett, Y., L. Herrera, J. Gonzalez, L. Cubilla, T. L. Capson, P. D. Coley, T. A. Kursar, L. I. Romero, and E. Ortega-Barria. 2004. A novel DNA-based microfluorimetric method to evaluate antimalarial drug activity. Am. J. Trop. Med. Hyg. 70:119-124. [PubMed] [Google Scholar]
- 11.Desjardins, R. E., C. J. Canfield, J. D. Haynes, and J. D. Chulay. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fidock, D. A., and E. H. Ekland. 2008. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int. J. Parasitol. 38:743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Happi, C. T., G. O. Gbotosho, A. Swowunmi, C. O. Falade, D. O. Akinboye, L. Gerena, D. E. Kyle, W. Milhous, D. F. Wirth, and A. M. J. Oduola. 2004. Molecular analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. Am. J. Trop. Med. Hyg. 70:20-26. [PubMed] [Google Scholar]
- 14.Happi, C. T., G. O. Gbotosho, O. A. Folarin, D. O. Akinboye, B. O. Yusuf, O. O. Ebong, A. Swowunmi, D. E. Kyle, W. Milhous, D. F. Wirth, and A. M. J. Oduola. 2005. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 95:183-193. [DOI] [PubMed] [Google Scholar]
- 15.Hastings, I. M., and U. D' Alessandro. 2000. Modelling a predictable disaster: the rise and spread of drug-resistant malaria. Parasitol. Today 16:340-347. [DOI] [PubMed] [Google Scholar]
- 16.Hyde, J. E. 2002. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 4:165-174. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. D., R. A. Dennull, L. Gerena, M. Lopez-Sanchez, N. E. Roncal, and N. C. Waters. 2007. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 51:1926-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen, J. H., and M. J. Ferraro. 2000. Antimicrobial susceptibility testing: special needs for fastidious organisms and difficult-to-detect resistance mechanisms. Clin. Infect. Dis. 30:799-808. [DOI] [PubMed] [Google Scholar]
- 19.Kosaisavee, V., R. Suwanarusk, F. Nosten, D. E. Kyle, M. Barrends, J. Jones, R. Price, B. Russell, and U. Lek-Uthai. 2006. Plasmodium vivax: isotopic, PicoGreen, and microscopic assays for measuring chloroquine sensitivity in fresh and cryopreserved isolates. Exp. Parasitol. 114:34-39. [DOI] [PubMed] [Google Scholar]
- 20.Liu, S., J. Mu, H. Jiang, and X.-Z. Su. 2008. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am. J. Trop. Med. Hyg. 79:178-184. [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzetti, A., P. A. Fornazari, A. C. Bonini-Domingos, R. D. S. R. Penhalbel, É. Fugikaha, C. R. Bonini-Domingos, V. D. Fraga, L. M. Conceição, A. R. B. Rossit, C. E. Cavasini, V. S. C. D. Couto, and R. L. D. Machado. 2008. Mixed Plasmodium faciparum infections and its clinical implications in four areas of the Brazilian Amazon region. Acta Trop. 107:8-12. [DOI] [PubMed] [Google Scholar]
- 22.Luxemburger, C., N. J. White, F. ter Kuile, H. M. Singh, I. Allier-Frachon, M. Ohn, T. Chongsuphajaisiddhi, and F. Nosten. 1996. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans. R. Soc. Trop. Med. Hyg. 90:105-111. [DOI] [PubMed] [Google Scholar]
- 23.May, J., and C. G. Meyer. 2003. Chemoresistance in falciparum malaria. Trends Parasitol. 19:432-435. [DOI] [PubMed] [Google Scholar]
- 24.Mayxay, M., S. Pukrittayakamee, P. N. Newton, and N. J. White. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20:233-240. [DOI] [PubMed] [Google Scholar]
- 25.Mbaisi, A., P. Liyala, F. Eyase, R. Achilla, H. Akala, J. Wangui, J. Mwangi, F. Osuna, U. Alam, B. L. Smoak, J. M. Davis, D. E. Kyle, R. L. Coldren, C. Mason, and N. C. Waters. 2004. Drug susceptibility and genetic evaluation of Plasmodium falciparum isolates obtained in four distinct geographical regions of Kenya. Antimicrob. Agents Chemother. 48:3598-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milhous, W. K., N. F. Weatherly, J. H. Bowdre, and R. E. Desjardins. 1985. In vitro activities of and mechanisms of resistance to antifol antimalarial drugs. Antimicrob. Agents Chemother. 27:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molineaux, L., J. Storey, J. E. Cohen, and A. Thomas. 1980. A longitudinal study of human malaria in the west African savanna in the absence of control measures: relationships between different Plasmodium species; in particular Plasmodium falciparum and Plasmodium malariae. Am. J. Trop. Med. Hyg. 29:725-737. [DOI] [PubMed] [Google Scholar]
- 28.Nakornchai, S., and P. Konthiang. 2006. Activity of azithromycin or erythromycin in combination with antimalarial drugs against multidrug-resistant Plasmodium falciparum in vitro. Acta Trop. 100:185-191. [DOI] [PubMed] [Google Scholar]
- 29.Oduola, A. M., N. F. Weatherly, J. H. Bowdre, and R. E. Desjardins. 1988. Plasmodium falciparum: cloning by single-erythrocyte micromanipulation and heterogeneity in vitro. Exp. Parasitol. 66:86-95. [DOI] [PubMed] [Google Scholar]
- 30.Ohrt, C., G. D. Willingmyre, P. Lee, C. Knirsch, and W. Milhous. 2002. Assessment of azithromycin in combination with other antimalarial drugs against Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 46:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plouffe, D., A. Brinker, C. McNamara, K. Henson, N. Kato, K. Kuhen, A. Nagle, F. Adrián, J. T. Matzen, P. Anderson, T. G. Nam, N. S. Gray, A. Chatterjee, J. Janes, S. F. Yan, R. Trager, J. S. Caldwell, P. G. Schultz, Y. Zhou, and E. A. Winzeler. 2008. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl. Acad. Sci. USA 105:9059-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postigo, M., A. Mendoza-León, and H. A. Pérez. 1998. Malaria diagnosis by the polymerase chain reaction: a field study in south-eastern Venezuela. Trans. R. Soc. Trop. Med. Hyg. 92:509-511. [DOI] [PubMed] [Google Scholar]
- 33.Rason, M. A., T. Radriantsoa, H. Andrianatenaina, A. Ratsimbasoa, and D. Menard. 2008. Performance and reliability of the SYBR green I based assay for the routine monitoring of susceptibility of Plasmodium falciparum clinical isolates. Trans. R. Soc. Trop. Med. Hyg. 102:346-351. [DOI] [PubMed] [Google Scholar]
- 34.Richie, T. L. 1988. Interactions between malarial parasites infecting the same vertebrate host. Parasitology 96:607-639. [DOI] [PubMed] [Google Scholar]
- 35.Sanders, J. W., G. S. Fuhrer, M. D. Johnson, and M. S. Riddle. 2008. The epidemiological transition: the current status of infectious diseases in the developed world versus the developing world. Sci. Prog. 91:1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smilkstein, M., N. Sriwilaijaroen, J. X. Kelly, P. Wilairat, and M. Riscoe. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial screening. Antimicrob. Agents Chemother. 48:1803-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ursos, L. M., and P. D. Roepe. 2002. Chloroquine resistance in the malarial parasite, Plasmodium falciparum. Med. Res. Rev. 22:465-491. [DOI] [PubMed] [Google Scholar]
- 38.Weisman, J. L., A. P. Liou, A. A. Shelat, F. E. Cohen, R. K. Guy, and J. L. DeRisi. 2006. Searching for new antimalarial therapeutics amongst known drugs. Chem. Biol. Drug Des. 67:409-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO. 2008. World malaria report 2008. World Health Organization, Geneva, Switzerland.
- 40.Willet, G. P., W. K. Milhous, L. Gerena, and A. M. J. Oduola. 1991. Mixed population dynamics in human malaria parasite cultures. Trans. R. Soc. Trop. Med. Hyg. 85:33-34. [DOI] [PubMed] [Google Scholar]
- 41.Wongsrichanalai, C., and S. R. Meshnick. 2008. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg. Infect. Dis. 14:716-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerman, P. A., R. K. Mehlotra, L. J. Kasehagen, and J. W. Kazura. 2004. Why do we need to know more about mixed Plasmodium species infections in humans? Trends Parasitol. 20:440-447. [DOI] [PMC free article] [PubMed] [Google Scholar]