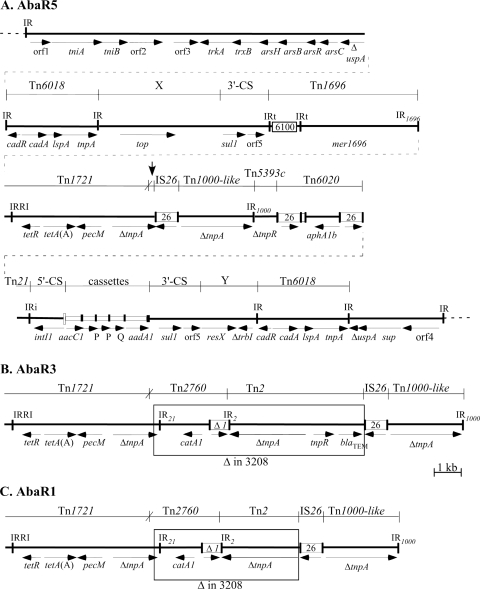
FIG. 1.
Map of AbaR5, showing the origins of different segments. (A) AbaR5. In the central line, the dotted lines represent chromosomal DNA flanking AbaR5, and IR of transposons are indicated with vertical lines. IR within the MARR between the Tn6018 regions have an identifying notation, IRi and IRt for class 1 integron ends and IR1696, IR21, IR2, and IR1000 for IR from Tn1696, Tn21, Tn2, and Tn1000-like, respectively. Numbered open boxes represent the IS, with the number indicating the identity of the IS. The attI1 site of the class 1 integron is shown as a tall open box, and cassettes are represented by an open box with a vertical bar representing the attC site. The lines above indicate the extents of the regions derived from known transposons, with vertical lines indicating the boundaries between segments and X indicating a recombination crossover. The mer region of Tn1696, encoding the merEDACTPR genes, is represented by mer1696. The extents of other genes are shown by horizontal arrows with the gene names underneath. The genes named resX, sup, and orf4 potentially encode a resolvase, a sulfate permease, and a protein of unknown function, respectively. The sup gene was incorrectly identified as sul1 in earlier publications (6, 20). The vertical arrow indicates the position of the deletion. (B and C) Portions of AbaR3 and AbaR1, respectively, that contain the catA1 and blaTEM genes. Only the region flanked by the segments from Tn1721 and Tn1000-like is shown, and the segment missing from AbaR5 is boxed. AbaR3 is otherwise identical to AbaR5, except that only one copy of the orfP cassette is present. AbaR1 contains a large segment replacing the region between Tn6018 and sul1, marked X in the in the top line of panel A. The structure of most of this region can be found in reference 8. Gene Construction Kit (version 2.5; Textco, West Lebanon, NH) was used to create the figure to scale.