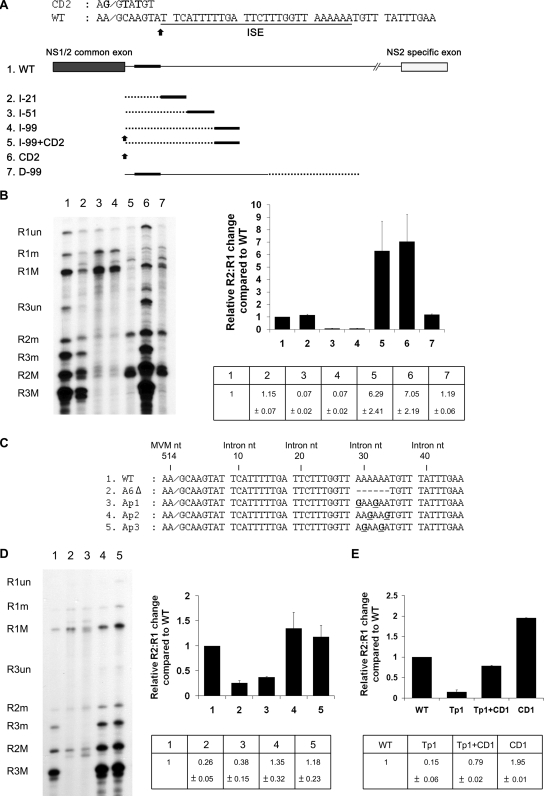
FIG. 2.
The activity of the ISE is distance dependent. (A) Top: Nucleotide sequence of the 5′ splice site region, indicating the position of the ISE and the insertion point for spacer additions. The sequence of the CD2 mutation is also shown (changes from the wild-type [WT] are in bold). Bottom: Diagram of insertion constructs described in the text. Dotted lines indicate heterologous spacer insertion fragments which were taken from a region of the capsid-coding gene of MVM not known to contain RNA-processing signals (see text). The black bars indicate the position of the ISE. Black arrows indicate consensus donor mutation CD2, which is described further in the text. The diagram is not to scale. (B) Left panel: Representative RNase protection analysis of constructs shown in panel A, using probe-spanning nt 1858 to 2377. Right panel: Quantification of RNase protection. The data, taken from at least three experiments, with standard deviations, are presented as the relative R2:R1 changes compared to that of the wild type in both tabular (bottom) and graphical (top) forms.