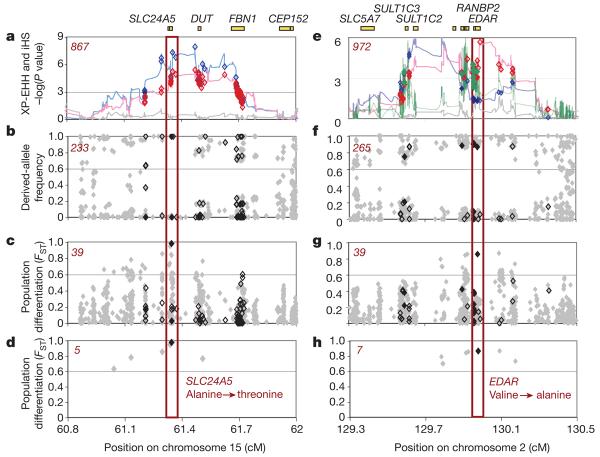
Figure 1. Localizing SLC24A5 and EDAR signals of selection.
a–d, SLC24A5. a, Strong evidence for positive selection in CEU samples at a chromosome 15 locus: XP-EHH between CEU and JPT + CHB (blue), CEU and YRI (red), and YRI and JPT + CHB (grey). SNPs are classified as having low probability (bordered diamonds) and high probability (filled diamonds) potential for function. SNPs were filtered to identify likely targets of selection on the basis of the frequency of derived alleles (b), differences between populations (c) and differences between populations for high-frequency derived alleles (less than 20% in non-selected populations) (d). The number of SNPs that passed each filter is given in the top left corner in red. The threonine to alanine candidate polymorphism in SLC24A5 is the clear outlier. e–h, EDAR. e, Similar evidence for positive selection in JPT + CHB at a chromosome 2 locus: XP-EHH between CEU and JPT + CHB (blue), between YRI and JPT + CHB (red), and between CEU and YRI (grey); iHS in JPT + CHB (green). A valine to alanine polymorphism in EDAR passes all filters: the frequency of derived alleles (f), differences between populations (g) and differences between populations for high-frequency derived alleles (less than 20% in non-selected populations) (h). Three other functional changes, a D→E change in SULT1C2 and two SNPs associated with RANBP2 expression (Methods), have also become common in the selected population.