Figure 3. Structural model of the EDAR death domain.
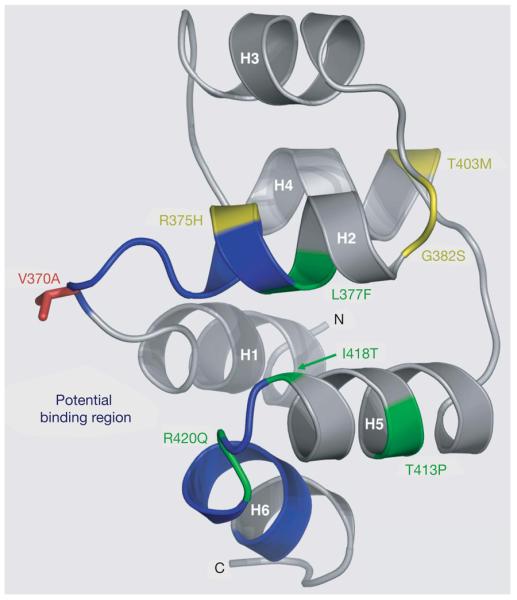
Ribbon representation of a homology model of the EDAR death domain (DD), based on the alignment of the EDAR DD amino acid sequence (EDAR residues 356–431), with multiple known DD structures. The helices are labelled H1 to H6. Residues in blue (the H1–H2 and H5–H6 loops, residues 370–376 and 419–425, respectively) correspond to the homologous residues in Tube that interact with Pelle in the Tube-DD–Pelle-DD structure24. These EDAR-DD residues therefore form a potential region of interaction with a DD-containing EDAR-interacting protein, such as EDARADD. The V370A polymorphic residue (red) is located prominently within this potential binding region in the H1–H2 loop. Seven of the thirteen known mis-sense mutations in EDAR that lead to hypohidrotic ectodermal dysplasia (HED) in humans are located in the EDAR-DD: the only four mutations in EDAR that lead to the dominant transmission of HED (green) and three recessive mutations (yellow)21. Four of these mutations, R375H, L377F, R420Q and I418T are located in the vicinity of the predicted interaction interface.
