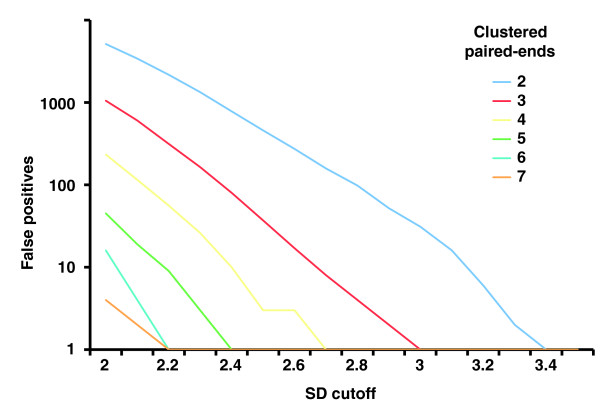
Figure 3.
Numbers of false positive SV calls in relation to the cutoff used for defining outliers. Cutoff values for defining outlier paired ends are given in terms of standard deviations (SDs) from the median of the expected distribution of paired-end spans (which in turn is derived from the insert size). PEM data generated with the 454/Roche platform were simulated applying a median insert size L = 2.5 kb and a span-coverage of λ = 5× of the diploid chromosome 2. To arrive at λ = 5×, only optimally (uniquely) placed paired ends were considered when estimating λ ('effective span coverage'). Here, the genome-wide count of false positives is put in relation to outlier-identification cutoffs for various required cluster sizes N ('clustered paired ends') of 2 up to 7. 'False positives' refers to the number of false positives identified on chromosome 2.