Short abstract
Where information on homologous proteins is available, progress is being made in automated prediction of protein function from sequence and structure.
Abstract
With many genomes now sequenced, computational annotation methods to characterize genes and proteins from their sequence are increasingly important. The BioSapiens Network has developed tools to address all stages of this process, and here we review progress in the automated prediction of protein function based on protein sequence and structure.
In the past, a protein's structure was usually experimentally determined after its biological role had been thoroughly elucidated, and the structure was used as a framework to explain known functional properties. This led to the view that the reliable prediction of the structure of a protein from its sequence would almost automatically provide information about its function. The good news is that methods for predicting structure from sequence can now produce good models for a substantial fraction of the protein space [1]. But the idea that knowledge of a protein's structure is sufficient for functional assignment has needed revision [2]. Many proteins of known structure are not yet functionally characterized and their number is increasing. The investigation of sequence-function and structure-function relationships has therefore become a fundamental necessity. Understanding these relationships will be crucial for moving from an inventory of protein parts to a more profound understanding of the molecular machinery of organisms at a systems level.
This review describes progress in developing both sequence-and structure-based methods for function prediction. There are many current methods, which use a variety of different approaches (Figure 1), and their integration is a major challenge. One approach to this problem has been employed by the BioSapiens Network [3] (see Box 1), of which we are members, through which several new methods have been developed and predictions integrated using the Distributed Annotation System (DAS) [4] (see Box 1). DAS allows different laboratories to 'combine' their annotations, produced by both experimental and computational approaches, to generate a 'composite' annotation at all levels. A 'protein sequence ontology' to facilitate comparison of the annotations has also been developed (G Reeves, personal communication).
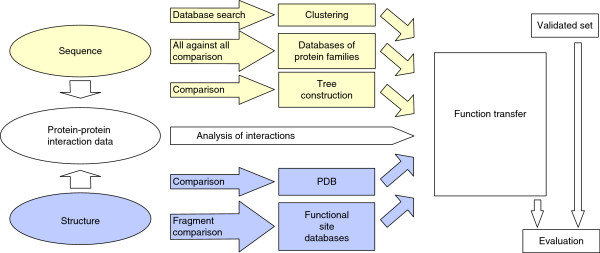
Figure 1.
Automated strategy for assigning function to proteins. The various approaches to protein function prediction are described in the text. Both protein sequences and structures can provide information for family classification and functional inference. Sequence-based methods make use of different strategies for grouping proteins into families (for example, sequence tree construction based on clustering of all against all sequence comparisons) or they compare the target sequence with pre-compiled databases of families. When a structure is available, the whole structure can be scanned against precompiled sets of functional sites. Alternatively, fragments of the target protein can be used to identify any structural similarities in the conformation of proteins of known structure, possibly related to a molecular function. Both sequences and structures, together with protein-protein interaction data, can be used to infer interactions, which can provide functional clues. Ideally, an independent set should be used to assess the reliability of the various methods.
Box 1.
Glossary of terms
We will first focus on methods that attempt to extract functional information from protein sequences, which generally exploit the power of alignment and clustering, and then discuss strategies that use protein structure information. When an experimental three-dimensional (3D) structure is not available, such methods can, in principle, be applied to modeled structures, although the quality of the model will dictate which methods can be applied (for a review, see [5]). We will also briefly discuss tools that exploit interactions between proteins as a means of inferring their function and survey systematic assessments of the effectiveness of function-prediction methods.
Sequence-based classification of proteins
The first hurdle for any functional annotation process is to define 'function'. If the protein is an enzyme, then simply using the EC numbering scheme (see Box 1) can be useful. In general however, the problem is multi-dimensional: a protein can have a molecular function, a cellular role, and be part of a functional complex or pathway (these are the distinctions used in the Gene Ontology (GO; see Box 1) [6]). Furthermore, certain aspects of molecular function can be illustrated by multiple descriptive levels (for example, the coarse 'enzyme' category versus a more specific 'protease' assignment). Even the more detailed definition would not reveal the cellular role of the protein (apoptosis, metabolism, blood coagulation, and so on).
Most function-prediction methods, both sequence and structure based, rely on inferring relationships between proteins that permit the transfer of functional annotations and binding specificities from one to the other. A notable challenge here is deciphering the connection between the detected similarities (structural or in sequence) and the actual level of functional relatedness. Function is often associated with domains, and another problem is the identification of functional domains from sequence alone. The accuracy of current methods for predicting domain boundaries is not yet completely satisfactory. Several methods provide reliable predictions if a structural template for the protein is available, but when this is not the case, one is left with the problem of whether the experimental annotation used for the inference refers to the same domain for which the sequence similarity/motif is established [7].
The function of a protein can also be inferred from its evolutionary relationship with proteins of known function, provided that the relationship is properly inspected. Orthologous proteins in different species most often share function, but paralogy (that is, divergence following duplication of the original gene) does not guarantee common function. Distinguishing between orthology and paralogy can be attempted on the basis of observed sequence-similarity patterns, by analyzing the specific conservation pattern of residues responsible for function in the family, or on the basis of the protein structure (either experimentally determined or modeled). In all cases, this requires the clustering of proteins into evolutionary families, which can be achieved using similarity-detection tools such as BLAST [8] or profiling tools based on multiple sequence alignments, for example, PSI-BLAST [9]. Several available resources provide pre-compiled family assignments for proteins on a genomic scale, based only on their sequence. Resources can be subdivided into those that consider full-length sequences and those based on domains or motifs that map to certain sub-sequences. In both cases, the degree of granularity of the classification is important, as this is related to the level of functional features that a group of proteins is expected to share.
A resource that classifies full-length proteins is PIRSF [10], in which a set of rules is applied to define primary and curated clusters that are also based on textual (protein names, literature) and parent-child relationships. These clusters (named superfamilies) are further divided into those with full-length similarity (that is, common domain architecture) and those sharing an ancestral domain. PIRSF covers more than two-thirds of the protein sequence space.
Studying proteins at a domain level allows more accurate functional inference [11] and is useful for predicting the function of novel domain combinations that possibly give rise to new protein functions [12]. In this type of resource, a family of domains is represented as a multiple sequence alignment, which is embodied in a statistical family signature profile (for example, CDD [13] and PROSITE [14]) or a profile-hidden Markov model (for example, Pfam [15] and SMART [16]), collectively referred to here as profiles. Pfam, a prototype for such collections, currently contains more than 9,000 family profiles and covers roughly 70-74% of UniProt sequences, capturing about half of their amino acids [17]. About 40-45% of Pfam families are associated with known structures, whereas 20-25% are currently uncharacterized. Other resources, for example CDD, use externally defined profiles to provide rapid assignments to sequence queries, using a BLAST-like engine to speed up searches.
Profile-based methods and resources differ significantly in their level of automation, their degree of manual curation, and the level of independence from complementary resources used in the classification. Combination of these resources provides a more comprehensive coverage, as reflected by InterPro [18], a repository of protein families integrating signatures from more than 10 member resources, currently covering nearly 75% of UniProt sequences. InterPro also includes Gene3d [19] and SUPERFAMILY [20], which provide sequence profiles corresponding to the structural classification of folds by CATH [21] and SCOP [22], respectively. A resource exploiting the multiplicity of essentially complete genome sequences is COG (Clusters of Orthologous Groups), an evolutionary classification that uses comparative genomics principles, such as phyletic profiles [23] (see Box 1), to identify the presence of orthologs, and group them accordingly.
A notable shortcoming of the methods described above is that they require definition of a threshold similarity for separating families from each other. An alternative approach to defining clusters is the construction of a tree representation that can provide a hierarchical view. Resources in this category include ProtoNet [24], CluSTr [25] and SYSTERS [26]. They are based on sequence similarities detected by an all-against-all sequence comparison, so that any level of evolutionary granularity can be inspected, from closely related subfamilies to more distant relationships.
Approaches that do not rely solely on supervised annotation of family profiles include ProDom [27], which collects putative domain profiles using known sequence domains as query sequences for iterative PSI-BLAST searches [9]. EVEREST [28] is a fully automatic unsupervised method that identifies recurrent conserved regions on the basis of local sequence similarities and iterative profile searches.
The accuracy of sequence-based methods is affected by the type and amount of information on the specific protein family but, overall, they seem to be reasonably accurate. Their success rate has been shown to be greater than 70% when tested on a limited dataset (all structures solved by the Midwest Center for Structural Genomics during the first five years of the Protein Structure Initiative) [29].
Structure-based methods
As homologous proteins evolve, their 3D structure often remains more conserved than their sequence [30]. Consequently, similarities in protein structure can be more reliable than sequence similarities for grouping together distant homologs, which often retain some aspect of a common biological function [31]. The two most comprehensive structure-based family resources, CATH [21] and SCOP [22], classify domains into evolutionary families and into coarser structural classes. Although both resources use some automated protocols, domain assignments are primarily made by expert manual validation. CATH differs from SCOP only in its use of structure-comparison algorithms (for example, CATHEDRAL [32]) and of hidden Markov model-based approaches to provide guidance to curators during classification (see Box 1). Creating functional subfamilies within superfamilies in CATH or SCOP not only permits the analysis of functional divergence with respect to structure, but can be used as a basis for structure-based function prediction. SCOP provides a level below superfamily that groups together closer homologs, often with more similar functions, and work is currently under way to offer similar information in CATH.
The first step in predicting function from structure is often to use global structure comparison: that is, to compare a query protein structure to domains in the structure databases. Although not directly coupled to a curated domain-family resource such as CATH and SCOP, other global structure-comparison methods (for example, DALI [33], MSDFold [34], VAST [35], CE [36], STRUCTAL [37] and FATCAT [38]) can identify structural neighbors in the Protein Data Bank (PDB) [39] (see Box 1), which may share functional similarities. Regardless of the algorithm used, care must be taken when transferring function from one protein to another, as two proteins may have a similar fold yet different functions (for example, the TIM-barrel scaffold).
Some algorithms exploit data on structural families to improve function prediction. The GASP method [40] applies a genetic algorithm to build templates made up of conserved residues in a given family of structures, which are evaluated on their ability to recognize other family members against a background of SCOP domains, when scanned using SPASM [41]. The DRESPAT [42] algorithm also identifies patterns within a family of proteins. The resulting structural motifs can be used to identify binding sites and to assign function to new structures.
Global structure-comparison methods are a useful first step for function assignment, but they do not discriminate between conservation of the overall fold and of functionally relevant regions of the protein. Other methods focus on more localized regions that might be relevant to function, such as clefts, pockets and surfaces. As the ligand-binding site or active site is commonly situated in the largest cleft in the protein [43], the identification and comparison of such regions can suggest putative functions. SURFNET [44] detects clefts by fitting spheres of a range of sizes between the protein's atoms and this approach has been enhanced by combining SURFNet with ConSurf [45] to identify only clefts that are close to evolutionarily conserved residues, as defined by the ConSurf-HSSP database [46]. Another surface-comparison method, pvSOAR [47], identifies similar surface patterns on the basis of geometrically defined pockets and voids. This approach and the associated CASTp [48] database have been used to create the Global Protein Surface Survey (GPSS). Functionally relevant surfaces (binding ligands, metals, DNA or peptide) are extracted through generation of an exclusion contact surface obtained by measuring the difference in solvent accessibility between a structure with and without a neighboring molecule.
Other pocket-centric approaches use the physicochemical properties of the local environments in the pockets and surfaces to describe protein-ligand interactions and active-site chemistry. For example, FEATURE [49] represents local microenvironment using various physical and chemical properties from atomic or chemical groups, from single residues up to secondary structure. Similar approaches are those of SiteEngine [50] and the recently released SURF'S UP! service [51].
Other methods target specific active-site residues (such as catalytic clusters and ligand-binding sites). These approaches utilize a variety of template-based scans to identify active sites and putative ligand-binding sites, the rationale being that the 3D arrangement of enzyme active-site residues is often more conserved than the overall fold. Templates can be derived manually by mining the literature and assessing which residues form the active site (for example, the Catalytic Site Atlas [52]), or can be generated automatically, as in PDBSiteScan [53,54], which uses the SITE records in PDB files and protein-protein interaction data to generate its templates. The Catalytic Site Atlas has been automatically expanded to include homologs identified by PSI-BLAST [9] and a new webserver (Catalytic Site Search) allows users to query the database directly [55].
Conventional template-searching tools scan the structure of the uncharacterized protein against a database of templates. This idea has been turned on its head with the 'reverse template' approach (initially developed as part of the ProFunc server [56]), which fragments a query protein into many putative templates and scans each of them against the PDB to identify similarities. A stand-alone version, Tempura, has recently been released at the European Bioinformatics Institute [57]. A similar approach is used by the PINTS (Patterns In Non-homologous Tertiary Structures) server [58], which detects the largest common 3D arrangement of residues between any two structures, the assumption being that similar arrangements of residues might imply relatedness of function. The latest addition to automatic template generation uses the Evolutionary Trace (ET) approach [59]. ET uses phylogenetic trees to rank residues in a protein by their evolutionary importance and maps these onto the structure, the highest-ranking residues tending to cluster on the protein surface in functionally important sites. This approach has been developed to build an automated Evolutionary Trace Annotation (ETA) pipeline [60,61] to identify functional sites, extract representative 3D templates and search for relevant geometric matches in other structures. Other template-centric approaches include Fuzzy Functional Forms (FFFs) [62] and SPASM/RIGOR [41].
Each method has its pros and cons and no single method is always successful. As a result, metaservers (see Box 1) have been developed that aim to combine many services to provide a consensus view that can often help researchers to identify the most likely functional predictions. The ProFunc server [56] is one such resource. It utilizes many of the previously described sequence-based and structure-based methods to present a summary of the most likely functions, represented by GO terms, in an intuitive and well linked web interface. A detailed benchmarking of ProFunc on structural genomics targets showed that, for the most successful methods, functional clues could be derived for approximately 60% of target proteins, of which about 70% were confidently predicted [29].
Another metaserver, ProKnow [63], also combines information from sequence and structural approaches, including fold similarity (DALI [33]) and templates (RIGOR [41]), with functional links taken from the DIP database of protein interactions [64]. The ProKnow authors quantified the level of the assigned function by the ontology depth (from 1 = general to 9 = specific) and showed that they can reach 89% correct assignments at ontology depth 1 and 40% at depth 9, with 93% coverage of 1,507 distinct folded proteins.
Finally, although technically not a structure-based approach, JAFA (Joined Assembly of Function Annotations) [65], is a metaserver that queries several function-prediction servers with a protein sequence to return a summary of predicted GO terms.
Protein interactions
Protein interactions provide a natural context for describing how these molecules catalyze metabolic reactions, build molecular machines and transmit cellular signals. The availability of high-throughput interaction data has enabled the 'guilt by association' principle to be applied to elucidating protein function. The exploitation of observed or predicted physical interactions to assign function is, however, complicated not only by the generally low quality of high-throughput data [66] and the sparseness of reliable interaction datasets derived from literature [67], but also by the sheer size of the problem. Recent estimates indicate that around 50,000 interactions may exist in yeast and more than 300,000 in human [68]. From the available 3D structures of protein complexes, the existence of around 10,000 distinct interaction types, defined by the particular mutual arrangement of their constituent subunits, has been proposed [69].
The vast variety of molecular interactions can, however, be reduced to a limited number of recurrent domain-interaction types. The domain composition of a protein can thus give functional clues. The iPfam [70] and 3did [71] databases provide pre-computed structural information about interactions for Pfam domains. When no 3D structure is available, domain interactions can be inferred: by identifying domain pairs that are significantly overrepresented in interacting proteins [72]; by coevolutionary analysis; by identifying correlated mutations that preserve favorable physico-chemical properties of putative interaction interfaces [73]; or by detecting correlated phylogenetic distributions due to coevolution [74]. A correlated whole-genome phylogenetic distribution of different domains could also indicate that they interact with each other directly or at least share a functional role. Two new web resources - DIMA [75] and DOMINE [76] - integrate predicted and known domain interactions into comprehensive domain-interaction networks.
Blind evaluations
The well established Critical Assessment of Techniques for Protein Structure Prediction (CASP) project [1,77] (see Box 1) has set up an additional function-prediction category. This is inherently different from the CASP structure-prediction categories, because at the end of the experiment the function of the target protein is likely to remain unknown. Nevertheless, the community concurred that the effort was justified because of its importance. The results of the first run were rather disappointing [78]: only a few groups participated in the challenge; 3D-structure predictions were rarely used for function prediction; and assessment procedure was too complex. Notably, however, the function predictions submitted by the different groups often agreed, and a 'consensus prediction' could be derived. A reassessment of the experiment after more experimental evidence had accumulated [79] revealed that a consensus prediction could reach as high as 80% accuracy, although the sample was too small to substantiate the significance of this finding.
CASP has fostered the development of many other experiments, such as AFP [80], BioCreative [81], GeneFun [82] and MouseFunc [83], that exploit a range of predictive methods to make functional annotations. Most computational methods participating in the AFP experiment to identify ligand-binding sites relied on the use of sequence and structural information from related proteins, and fell into the broad categories described above [80]. Only rarely did predictors attempt to identify ligand-binding sites de novo. In the second edition of the AFP experiment, in 2007, some novel ideas were explored, such as the potential contribution of protein disorder, and a systems-level analysis of pathways.
Regretfully, the lack of independent test sets for testing blind predictions prevents proper assessment of the strengths and caveats of individual methods. This general issue should concern the whole biological community. It will be difficult to improve function-prediction methods without reliable test sets and a good way to overcome this problem has yet to be found.
In summary, function prediction remains a challenge. Ab initio prediction (that is, not based on annotation transfer) usually provides very limited, if any, clues. Evolutionary relationships, though complicated by the ortholog/paralog dichotomy, are by far the strongest predictors and the next few years will see increasingly sophisticated methods for deciphering their functional meaning. Molecular biology is moving to a more holistic view of biological processes and this requires better integration of different types of data. Elucidating function needs the combination of information from genomes, sequences, transcription patterns and genetic variation, as well as the results of prediction algorithms. Community approaches will ultimately empower discovery-oriented biology and, in turn, improve its translation to medicine and the environment.
Acknowledgments
Acknowledgements
This work was supported by the BioSapiens project, funded by the European Commission within its FP6 Programme, under the thematic area 'Life sciences, genomics and biotechnology for health', contract number LSHG-CT-2003-503265.
References
- Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69(Suppl 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Structural genomics: keeping up with expanding knowledge of the protein universe. Curr Opin Struct Biol. 2007;17:347–353. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GA, Thornton JM. Integrating biological data through the genome. Hum Mol Genet. 2006;15(Spec No 1):R81–R87. doi: 10.1093/hmg/ddl086. [DOI] [PubMed] [Google Scholar]
- Prlic A, Down TA, Kulesha E, Finn RD, Kahari A, Hubbard TJ. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontano A. The role of molecular modelling in biomedical research. FEBS Lett. 2006;580:2928–2934. doi: 10.1016/j.febslet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress M, Cheng J, Baldi P, Joo K, Lee J, Seo JH, Lee J, Baker D, Chivian D, Kim D, Ezkurdia I. Assessment of predictions submitted for the CASP7 domain prediction category. Proteins. 2007;69(Suppl 8):137–151. doi: 10.1002/prot.21675. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, Ledley RS, Suzek BE, Arminski L, Chen Y, Zhang J, Cardenas JL, Chung S, Castro-Alvear J, Dinkov G, Barker WC. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;(32 Database):D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AJ, Yeats C, Orengo CA. Methods of remote homology detection can be combined to increase coverage by 10% in the midnight zone. Bioinformatics. 2007;23:2353–2360. doi: 10.1093/bioinformatics/btm355. [DOI] [PubMed] [Google Scholar]
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;(35 Database):D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;(36 Database):D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;(36 Database):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;(34 Database):D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The universal protein resource (UniProt). Nucleic Acids Res. 2008;(36 Database):D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;(35 Database):D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats C, Lees J, Reid A, Kellam P, Martin N, Liu X, Orengo C. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;(36 Database):D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;(35 Database):D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH - a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/S0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Sasson O, Inbar U, Friedlich M, Fromer M, Fleischer H, Portugaly E, Linial N, Linial M. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;(33 Database):D216–D218. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Kretschmann E, Wieser D, Apweiler R. The predictive power of the CluSTr database. Bioinformatics. 2005;21:3604–3609. doi: 10.1093/bioinformatics/bti542. [DOI] [PubMed] [Google Scholar]
- Krause A, Stoye J, Vingron M. Large scale hierarchical clustering of protein sequences. BMC Bioinformatics. 2005;6:15. doi: 10.1186/1471-2105-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;(33 Database):D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugaly E, Linial N, Linial M. EVEREST: a collection of evolutionary conserved protein domains. Nucleic Acids Res. 2007;(35 Database):D241–D246. doi: 10.1093/nar/gkl850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Sanderson S, Ezersky A, Savchenko A, Edwards A, Orengo C, Joachimiak A, Laskowski RA, Thornton JM. Towards fully automated structure-based function prediction in structural genomics: a case study. J Mol Biol. 2007;367:1511–1522. doi: 10.1016/j.jmb.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Orengo CA. Protein structure alignment. J Mol Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- Redfern OC, Harrison A, Dallman T, Pearl FM, Orengo CA. CATHEDRAL: a fast and effective algorithm to predict folds and domain boundaries from multidomain protein structures. PLoS Comput Biol. 2007;3:e232. doi: 10.1371/journal.pcbi.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J Mol Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacco BJ, Babbitt PC. Automated discovery of 3D motifs for protein function annotation. Bioinformatics. 2006;22:723–730. doi: 10.1093/bioinformatics/btk038. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- Wangikar PP, Tendulkar AV, Ramya S, Mali DN, Sarawagi S. Functional sites in protein families uncovered via an objective and automated graph theoretic approach. J Mol Biol. 2003;326:955–978. doi: 10.1016/S0022-2836(02)01384-0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F, Rosenberg Y, Kessel A, Pupko T, Ben-Tal N. The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins. 2005;58:610–617. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- Binkowski TA, Freeman P, Liang J. pvSOAR: detecting similar surface patterns of pocket and void surfaces of amino acid residues on proteins. Nucleic Acids Res. 2004;32:W555–W558. doi: 10.1093/nar/gkh390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley SC, Altman RB. Characterizing the microenvironment surrounding protein sites. Protein Sci. 1995;4:622–635. doi: 10.1002/pro.5560040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ. SiteEngines: recognition and comparison of binding sites and protein-protein interfaces. Nucleic Acids Res. 2005;33:W337–W341. doi: 10.1093/nar/gki482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasin JM, Godzik A, Bujnicki JM. SURF'S UP! - protein classification by surface comparisons. J Biosci. 2007;32:97–100. doi: 10.1007/s12038-007-0009-0. [DOI] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSiteScan: a program for searching for active, binding and posttranslational modification sites in the 3D structures of proteins. Nucleic Acids Res. 2004;32:W549–W554. doi: 10.1093/nar/gkh439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33:D183–D187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA, Spriggs RV, Bartlett GJ, Gutteridge A, MacArthur MW, Porter CT, Al-Lazikani B, Thornton JM, Swindells MB. Effective function annotation through catalytic residue conservation. Proc Natl Acad Sci USA. 2005;102:12299–12304. doi: 10.1073/pnas.0504833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempura http://www.ebi.ac.uk/thornton-srv/databases/tempura
- Stark A, Russell RB. Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures. Nucleic Acids Res. 2003;31:3341–3344. doi: 10.1093/nar/gkg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Kristensen DM, Ward RM, Lisewski AM, Erdin S, Chen BY, Fofanov VY, Kimmel M, Kavraki LE, Lichtarge O. Prediction of enzyme function based on 3D templates of evolutionarily important amino acids. BMC Bioinformatics. 2008;9:17. doi: 10.1186/1471-2105-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RM, Erdin S, Tran TA, Kristensen DM, Lisewski AM, Lichtarge O. De-orphaning the structural proteome through reciprocal comparison of evolutionarily important structural features. PLoS ONE. 2008;3:e2136. doi: 10.1371/journal.pone.0002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgard S, Cammer SA, Hoffman BT, Knutson S, Gallina M, Speir JA, Fetrow JS, Baxter SM. Prediction of deleterious functional effects of amino acid mutations using a library of structure-based function descriptors. Proteins. 2003;53:806–816. doi: 10.1002/prot.10458. [DOI] [PubMed] [Google Scholar]
- Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg I, Harder T, Godzik A. JAFA: a protein function annotation meta-server. Nucleic Acids Res. 2006;34:W379–W381. doi: 10.1093/nar/gkl045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stümpflen V, Mewes HW, Ruepp A, Frishman D. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- Finn RD, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R, Lee C, Sabatti C, Eisenberg D. Inferring protein domain interactions from databases of interacting proteins. Genome Biol. 2005;6:R89. doi: 10.1186/gb-2005-6-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cherukuri PF, Tasneem A, Przytycka TM. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein-protein interactions. J Mol Biol. 2006;362:861–875. doi: 10.1016/j.jmb.2006.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel P, Wong P, Frishman D. A domain interaction map based on phylogenetic profiling. J Mol Biol. 2004;344:1331–1346. doi: 10.1016/j.jmb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Pagel P, Oesterheld M, Tovstukhina O, Strack N, Stumpflen V, Frishman D. DIMA 2.0--predicted and known domain interactions. Nucleic Acids Res. 2008;(36 Database):D651–D655. doi: 10.1093/nar/gkm996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari B, Tasneem A, Przytycka TM, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2008;(36 Database):D656–D661. doi: 10.1093/nar/gkm761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- Soro S, Tramontano A. The prediction of protein function at CASP6. Proteins. 2005;61(Suppl 7):201–213. doi: 10.1002/prot.20738. [DOI] [PubMed] [Google Scholar]
- Pellegrini-Calace M, Soro S, Tramontano A. Revisiting the prediction of protein function at CASP6. FEBS J. 2006;273:2977–2983. doi: 10.1111/j.1742-4658.2006.05309.x. [DOI] [PubMed] [Google Scholar]
- AFP http://biofunctionprediction.org
- BioCreative http://biocreative.sourceforge.net
- GeneFun http://www.genefun.org
- MouseFunc http://www.mousefunc.org