Figure 1.
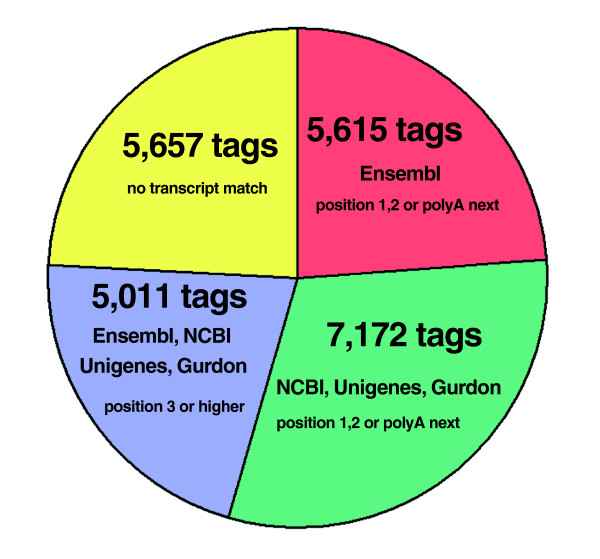
Tag-mapping of experimental tags to X. tropicalis genome and transcript databases. All different experimental tags (23,766 tags) were mapped first to the genome of X. tropicalis and those without a match (311 tags) were discarded from further analysis. The remaining experimental tags that presented one or more matches to the genome (23,455 tags; 100%) were then mapped to the Ensembl modified database, and only those tags found in the first or second positions from the 3'-end of the RNA sequence or belonging to the polyA-next category (see Materials and methods for details) were selected and reported as mapping to this transcript database (5,615 tags; 23.9%; red). The remaining tags that did not exhibit a match to the transcripts in the Ensembl modified database (17,840; 76.1%) were then searched with the same restraints mentioned above in the joint set composed of the NCBI (mRNAs), Unigene (clusters of mRNAs and ESTs) and Gurdon databases (clusters of ESTs). A total of 7,172 tags (30.6%) were found to match to positions 1, 2 or poly-A next in the transcripts from this set (green). The remaining tags without a match to these databases (10,668; 45.5%) were then re-mapped against the complete set of transcripts (a complete joint set of RNAs composed of Ensembl, NCBI, Unigene and Gurdon databases), but with the restraint that the mapping must occur to position 3 or above in a transcript. A total of 5,011 tags (21.4%) that fulfilled these conditions were obtained (blue). The remaining 5,657 (24.1%) tags mapped to the genome, but did not map to any known transcript (yellow).