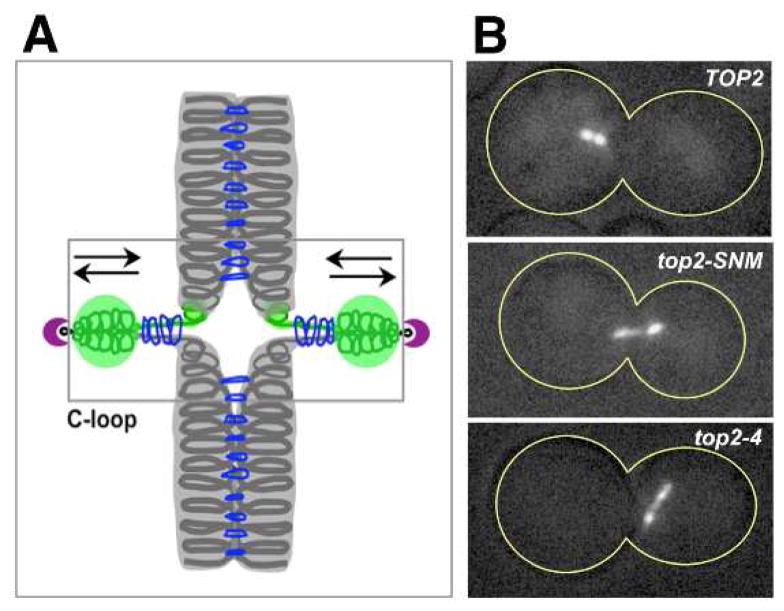
Figure 3. Influence of Top2 on budding yeast centromere dynamics.
(A) Diagram of proposed C-loop structure for mitotic CEN chromatin [187]. In this model, the separation of CEN chromatid fibers that occurs in response to bipolar spindle traction is stabilized by a localized reorganization of cohesin (blue rings) to form intra-stand cohesive linkages. GFP labeled chromatin (green; [188]) and kinetochores (purple) are also depicted. (B) Wild type (TOP2), top2–4 and SUMO conjugation resistant top2-SNM (see section 4.2.) strains were blocked in mitosis and the stretching of CEN chromatin under tension was evaluated using a CEN-proximal GFP tag on chromosome IV (CEN4-GFP). These images are reproduced here from [126] and from J. Bachant, InCytes, Mol. Biol. Cell 19 (2008) 4019 with permission from the publisher.