Abstract
Translation initiation in eukaryotes is accomplished by a large set of translation initiation factors, some of which are regulated by signals monitoring intracellular and environmental conditions. Here, we show that Uri1p is required for efficient translation initiation in budding yeast. Indeed, uri1Δ cells are slow growing, sensitive to translation inhibitors and they exhibit an increased 80S peak in polysome profiles. Moreover, GCN4 translation is derepressed in uri1Δ cells, strongly supporting an initiation defect. Genetic and biochemical experiments indicate that Uri1p interacts with the translation initiation factor eIF1A and promotes ternary complex (TC) recruitment to the 40S subunit. Interestingly, we found that Uri1p is also part of a chaperone-network, including the prefoldin Pfd6p and several other proteins involved in cotranslational quality control such as the ribosome-associated Hsp70 chaperone Ssb1p, the Hsp40 Sis1p and the translation elongation factor eEF1A. Together with genetic data, these interactions indicate that Uri1p may coordinate translation initiation and cotranslational quality control.
Keywords: chaperones, prefoldin, protein quality control, translation initiation, URI
Introduction
Regulated protein synthesis is a fundamental process that is carried out by ribosomes and a large number of translation factors. Chaperone complexes assist in the cotranslational folding of nascent polypeptides into their functional conformations (reviewed in Frydman, 2001). Protein translation can be divided into three successive events. First, translation initiation leads to the correct positioning of the 80S ribosome at the AUG start codon, which in eukaryotes is accomplished by a plethora of well-described initiation factors. The process begins with the recruitment of a ternary complex (TC) consisting of the initiator methionyl-tRNA (Met-tRNAiMet), the initiation factor eIF2 and GTP to the 40S ribosomal subunit. The initiation factor eIF1A promotes and stabilizes TC binding to the 40S subunit, resulting in the formation of the 43S preinitiation complex (PIC), which besides the 40S subunit and the TC contains many other associated factors. The PIC is then recruited to the mRNA through the eIF4F-initiation factors that are associated with the 5′cap structure of the mRNA. The resulting complex scans the mRNA for the AUG codon, and recognition of the start codon triggers GTP hydrolysis by eIF2 followed by release of the initiation factors and joining of the 60S subunit. These rearrangements result in an elongation-competent ribosome, which is able to begin synthesis of the polypeptide (reviewed in Dever, 2002; Gebauer and Hentze, 2004).
The second step of protein synthesis, translation elongation, ensures the processive addition of amino acids to the growing polypeptide. Elongation is followed by translation termination, which leads to the release of the protein at the stop codon. Nascent and newly synthesized proteins are targets of co- and post-translational quality control mechanisms. In eukaryotes, several sets of chaperones assist the folding of nascent chains, including the nascent chain-associated complex, the Hsp70 system, the TriC chaperonin system and prefoldins (PFDs) (reviewed in Frydman, 2001). Strikingly, it has been estimated that up to 50% of newly synthesized proteins are cotranslationally degraded by the proteasome (Schubert et al, 2000; Turner and Varshavsky, 2000), implying that efficient quality control mechanisms must exist that couple protein translation, protein folding and proteasomal degradation. Mistranslated and misfolded proteins may be detrimental for the cell and need to be removed at an early stage. However, to date, the mechanisms and coordination between translation and quality control remain poorly understood.
Protein synthesis is regulated in response to extracellular signals and intracellular growth and stress conditions, primarily at the level of translation initiation. Moreover, several chaperone systems are upregulated in response to stress conditions to cope with the increase of misfolded or denatured proteins. The two best-described mechanisms that control translation initiation include first, sequestration of the cap-binding eIF4-complex by eIF4E-binding proteins (4E-BPs), which is reversed under nutrient-rich conditions by phosphorylation of 4E-BP by the TORC1 kinase. The second initiation control mechanism involves Gcn2p-dependent phosphorylation of eIF2α, which converts it into an inhibitor of its own guanine-nucleotide exchange factor (GEF) eIF2B (Figure 1A) (reviewed in Dever, 2002; Gebauer and Hentze, 2004). Although the latter mechanism inhibits translation of most proteins, reduced TC levels are required to activate translation of a few stress-regulated proteins such as the transcription factors ATF4 in mammals and Gcn4p in yeast. Gcn4p controls the expression of >500 target genes, among them many involved in amino acid biosynthesis. Its protein levels are upregulated in response to amino acid deprivation and other types of starvation or stress conditions by an elaborate translation regulation mechanism involving four upstream open reading frames (uORFs) in the 5′UTR of GCN4. Under rich conditions, when TC concentration is high, scanning 40S subunits are efficiently reloaded with TC after uORF1 and restart translation at uORFs 2, 3 or 4. Because of the GC-rich sequences surrounding the uORF4 stop codon, 80S ribosomes disassemble and do not reform at the GCN4 start site, resulting in low levels of translated Gcn4p. Starvation conditions reduce TC levels, thereby causing less efficient TC re-loading after uORF1. Under these conditions, uORFs 2–4 are mostly by-passed and the PIC is not able to form before scanning has advanced to the GCN4 start codon, resulting in increased GCN4 translation under starvation conditions (reviewed in Hinnebusch, 2005).
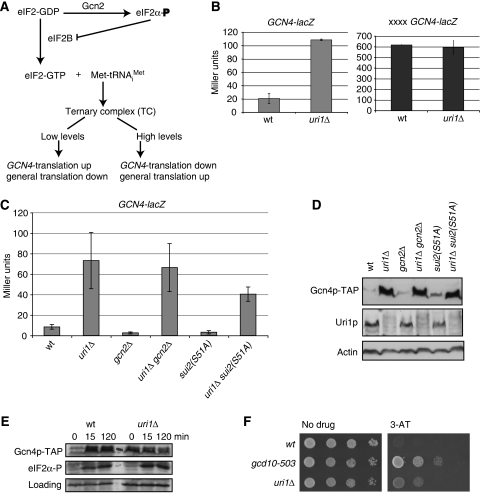
Figure 1.
Uri1p regulates GCN4 translation independently of eIF2α (Sui2p) phosphorylation. (A) Diagram representing regulation of GCN4 translation and general translation by ternary complex (TC) levels. (B, C) The indicated strains transformed with GCN4-lacZ (uORFs 1–4) (pBL199) or xxxxGCN4-lacZ (uORFs with mutated initiation codons) (pBL201) reporters were grown under rich conditions, and β-galactosidase activity in extracts was quantified in Miller units. Error bars represent standard deviations of three (B) or four (C) individual transformants. The following strains were analysed: wild-type (YAD219), uri1Δ (YAD211), gcn2Δ (YAD352), gcn2Δ uri1Δ (YAD310), sui2Δ (pSUI2(S51A)) (YAD23) and uri1Δ sui2Δ (pSUI2(S51A)) (YAD324). (D) Protein extracts of non-starved wild-type (YAD135), uri1Δ (YAD327), gcn2Δ (YAD353), gcn2Δ uri1Δ (YA355), sui2Δ (pSUI2(S51A)) (YAD359) and uri1Δ sui2Δ (pSUI2(S51A)) (YAD357) cells carrying TAP-tagged Gcn4p expressed from its genomic locus were blotted and probed with antibodies specific for Uri1p, proteinA (recognizing Gcn4p-TAP) and actin. (E) Exponentially growing wild-type (YAD135) and uri1Δ (YAD327) cells harbouring endogenously TAP-tagged Gcn4p were pelleted and resuspended in starvation medium lacking amino acids. TCA extracts were prepared at the indicated time points and the levels of Gcn4p-TAP and S51-phosphorylated eIFα was analysed by immunoblotting. A non-specific band serves as a loading control (bottom panel). (F) Serial dilutions of an equal number of his1-29 gcn2-101 gcn3-101 (YAD37), his1-29 gcn2-101 gcn3-101 gcd10-503 (YAD36) and his1-29 gcn2-101 gcn3-101 uri1::Nat (YAD379) cells were spotted on plates with complete SD medium or SD medium lacking histidin containing 10 mM 3-aminotriazole (3-AT). Plates were grown for 1–3 days at 30°C.
This regulated mechanism of GCN4 translation was successfully used as a genetic tool to identify factors involved in translation initiation. Indeed, GCN4 expression under rich conditions indicates defects in translation initiation and several so-called Gcd− mutations (general control derepressed) were identified where the amount of TC bound to the 40S subunit was reduced and therefore GCN4 translation was increased. Typical Gcd− mutants lead to GCN4 derepression even in the absence of functional Gcn2p, suggesting that they affect proteins that regulate GCN4 translation downstream or independently of Gcn2p (reviewed in Hinnebusch, 2005).
It has been shown earlier that GCN4 is constitutively expressed in yeast cells lacking URI1 (Unconventional prefoldin Rpb5 Interactor, also known as BUD27) (Gstaiger et al, 2003). On the basis of the observation that yeast uri1Δ cells exhibit starvation-like phenotypes and that hURI is regulated by the TOR pathway in mammalian cells, it has been suggested that Uri1p has a conserved function in nutritional signalling. The Uri1 protein contains evolutionary conserved Rpb5p-binding and PFD domains, which have been shown to mediate binding of Uri1p to Rpb5p and Pfd6p, respectively. PFD domains are typical for PFDs, which in the PFD/GimC complex are known to function as molecular chaperones that assist in the folding of newly synthesized actin and tubulin (Geissler et al, 1998; Siegers et al, 1999). However, Uri1p is not a subunit of the canonical GimC complex and so far no functional connection to other PFDs has been reported. Human URI forms a PFD/GimC-like complex together with the PFD-proteins STAP1, PFD2 and PFD4-related (PFD4r) as well as the non-PFDs RPB5, TIP48 and TIP49 (Gstaiger et al, 2003). Moreover, hURI was found to associate with PP1γ at mitochondria, where it is involved in a feedback loop to inhibit S6 kinase 1 on growth factor stimulation (Djouder et al, 2007). At present it is not clear whether these different human URI complexes are functionally connected.
In this study, we analysed the function of yeast Uri1p, starting from the observation that uri1Δ cells confer a Gcd− phenotype. We found that Uri1p is involved in translation initiation, where it promotes TC recruitment to the 40S subunit. Moreover, Uri1p interacts with several nascent chain-associated proteins, and deletion of URI1 renders chaperone mutants resistant to the misfolding inducing drug azetidine-2-carboxylic acid (AZC). Thus, we propose that Uri1p may be involved in coordinating translation initiation with cotranslational quality control.
Results
Uri1p is required to repress GCN4 translation independently of eIF2α-(Sui2p) phosphorylation
Earlier studies have shown that Gcn4p is upregulated in uri1Δ cells by a mechanism that involves the uORFs in the 5′UTR of GCN4 (Gstaiger et al, 2003). To investigate the molecular basis of this regulation, we compared the expression of wild type (GCN4-lacZ) and GCN4-lacZ reporter constructs harbouring mutations in the initiation codons of all four upstream uORFs (xxxxGCN4-lacZ) (Mueller and Hinnebusch, 1986). GCN4-lacZ expression was repressed in wild type but not uri1Δ cells (Figure 1B), whereas the xxxxGCN4-lacZ reporter was expressed at similar levels in both strains. These data imply that GCN4 derepression in uri1Δ cells is not dependent on the promoter region, which is shared by both constructs, but requires functional uORFs.
A well-known mechanism to upregulate GCN4 translation involves Gcn2p-dependent phosphorylation of eIF2α on Ser51 (Figure 1A). However, deletion of URI1 leads to a strong GCN4-lacZ induction in gcn2Δ cells, indicating that Uri1p function is not dependent on Gcn2p (Figure 1C; Gstaiger et al, 2003). Moreover, GCN4-lacZ expression was also induced in uri1Δ cells expressing a non-phosphorylatable S51A mutant of eIF2α (encoded by SUI2) (Figure 1C), excluding the possibility that a kinase other then Gcn2p may phosphorylate Ser51 under these conditions. Immunoblotting experiments confirmed that TAP-tagged Gcn4p is expressed in uri1Δ, uri1Δ gcn2Δ and uri1Δ sui2(S51A) cells grown in nutrient-rich conditions (Figure 1D), whereas it was barely detectable in wild type, gcn2Δ and sui2(S51A) cells. We could not detect increased levels of eIF2α phosphorylation in a uri1Δ strain under non-starvation conditions, whereas phosphorylation was efficiently induced on amino acid starvation in both wild-type and uri1Δ cells (Figure 1E).
Gcn2p-independent derepression of GCN4 translation under rich conditions is typical for Gcd− mutants (Harashima and Hinnebusch, 1986). Originally, Gcd− mutants were isolated as suppressors of the inability of gcn2-101 gcn3-101 double mutants to activate GCN4 and its target genes in the histidine biosynthetic pathway. In a his1-29 background, gcn2-101 gcn3-101 double mutants are hypersensitive to 3-amino triazole (3-AT), an inhibitor of histidine biosynthesis, and Gcd− mutants can suppress this sensitivity (Harashima and Hinnebusch, 1986). Indeed, deletion of the URI1 gene in a gcn2-101 gcn3-101 his1-29 strain restored its ability to grow on 3-AT-containing media, similarly to gcd10-503 controls (Figure 1F). Taken together, these results show that, similar to known Gcd-proteins, Uri1p is required to repress GCN4 translation under nutrient-rich conditions.
Cells lacking Uri1p are slow growing, hypersensitive to translation inhibitors and exhibit a translation initiation defect
Defects in protein translation generally cause reduced fitness such as slow growth and temperature sensitivity (Cuesta et al, 1998). Consistent with a role for Uri1p in protein translation, uri1Δ cells divided with a doubling time of 2.34 h compared with 1.52 h for isogenic wild-type controls (Figure 2A). Moreover, uri1Δ cells display reduced growth at 37°C and were hypersensitive to the translation inhibitors cycloheximide, paramomycin and hygromycin B (Figure 2B, data not shown). In contrast, uri1Δ cells were not hypersensitive to the DNA damaging agent methyl methan sulfonate, indicating that the sensitivity to translation inhibitors is specific and not caused by a general uptake defect (Supplementary Figure 1). All these sensitivities could be rescued by transforming uri1Δ cells with a plasmid carrying the URI1 gene. As Gcn4p levels are induced in uri1Δ cells leading to upregulation of Gcn4p targets such as Gln3p and Gat1p (Gstaiger et al, 2003), we analysed the phenotype of gcn4Δ uri1Δ double mutants. GCN4 deletion was unable to rescue the sensitivities and slow growth defects of uri1Δ cells (Figure 2A and B), indicating that these defects are not caused by upregulated Gcn4p targets.
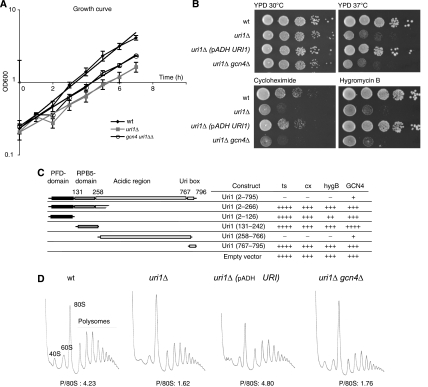
Figure 2.
Deletion of URI1 affects general translation. (A) Over night cultures of wild-type (YAD219), uri1Δ (YAD211) and uri1Δ gcn4Δ (YAD308) strains were diluted to OD600 0.2 and grown in YPD for 9 h. OD600 was measured at the indicated time points, and plotted against time (in hours) in semi-logarithmic graphical form. Bold lines represent exponential trend lines. Error bars represent the standard deviation of three independent experiments. Growth rates are wild-type, 1.5 h; uri1Δ, 2.34 h; uri1Δ gcn4Δ, 2.15 h. (B) Wild-type (YAD219), uri1Δ (YAD211) and gcn4Δ uri1Δ (YAD308) strains were transformed with an empty control vector or a plasmid expressing Uri1p from the ADH-promoter (pAD89). An equal number of cells were spotted in serial dilutions on YPD plates containing the indicated drugs in the following concentrations: cycloheximide 0.1 μg/ml, hygromycin B 0.05 mM. YPD plates without drugs were incubated for 2 days at 30 and 37°C, respectively, strains on drug plates were grown at 30°C for 3 days. (C) uri1Δ cells (YAD213) were transformed with plasmids expressing the indicated Uri1p fragments from the ADH-promoter, and tested for their ability to grow at 37°C (ts) or on plates containing the indicated drugs. The transformants were also tested for their ability to repress HIS4-LacZ expression (GCN4). In the lacZ-assay, increasing numbers of + indicate higher GCN4 expression, whereas for the sensitivity assays, more + indicate increased sensitivity. cx, cycloheximide; hygB, hygromycin B; GCN4, HIS4-lacZ assay. (D) Representative polysome traces of wild-type (YAD219), uri1Δ (YAD211) and gcn4Δ uri1Δ (YAD308) cells carrying an empty vector (pAD49), and uri1Δ cells transformed with a plasmid expressing URI1 from the ADH-promoter (pAD89). Extracts were prepared as described in Materials and methods. Samples were centrifuged in low salt (100 mM NaCl) buffer on 15 to 50% sucrose gradients, and polysome profiles were obtained by measuring absorption at 254 nm. Polysome/80S ratios are indicated.
To test which conserved domains of Uri1p are involved in the uncovered translation function of Uri1p, we transformed uri1Δ cells with plasmids expressing full-length Uri1p or the indicated Uri1p domains, and tested their ability to complement the temperature sensitivity, GCN4 derepression and sensitivity to translation inhibitors (Figure 2C). Surprisingly, neither the PFD nor the Rpb5p-binding domain (RPB5) were functionally important in vivo, whereas the acidic region of Uri1p was necessary and sufficient to rescue all tested translation defects of uri1Δ cells.
To directly assess the role of Uri1p in protein synthesis, we analysed the proportion of actively translating ribosomes in polysome profiles of wild-type and uri1Δ cells. Yeast cell extracts were centrifuged through sucrose gradients to separate free ribosomal subunits from 80S ribosomes and polysomes. Interestingly, the 80S peak was considerably higher in extracts prepared form uri1Δ cells compared with wild-type controls, and the corresponding polysomes/80S ratio was significantly reduced (Figure 2D). A wild-type polysome profile was restored after introducing a plasmid carrying URI1, showing that this translation defect is indeed caused by the absence of Uri1p. Consistent with the results described above, deletion of the GCN4 gene in uri1Δ cells did not suppress the translation effect analysed by polysome gradients. Taken together, we conclude that Uri1p is required for efficient protein translation, by a mechanism independent of Gcn4p.
Uri1p regulates translation initiation by promoting efficient recruitment of the TC to 40S subunits
The observed translation defects in uri1Δ cells support a role of Uri1p in translation initiation, possibly by promoting recruitment of the TC to the 40S ribosome. We thus tested whether the Gcd– phenotype of uri1Δ cells could be suppressed by a multicopy plasmid (hcTC) overexpressing all components of the TC, consisting of three eIF2 subunits and Met-tRNAiMet (Asano et al, 1999). GCN4 expression was measured indirectly by following the activity of its target gene HIS4, either using a specific reporter construct (HIS4-LacZ), where Gcn4p drives the expression of β-galactosidase (Figure 3A), or the sensitivity to the histidine-biosynthesis inhibitor 3-AT (Figure 3B). Interestingly, TC overexpression reduced HIS4-lacZ levels in uri1Δ strains almost three-fold (Figure 3A), and likewise, TC overexpression rescued the 3-AT-resistence of uri1Δ cells (Figure 3B). Together, these results provide genetic evidence that Uri1p may promote TC recruitment to 40S ribosomes (as suggested for eIF1A in Olsen et al, 2003).
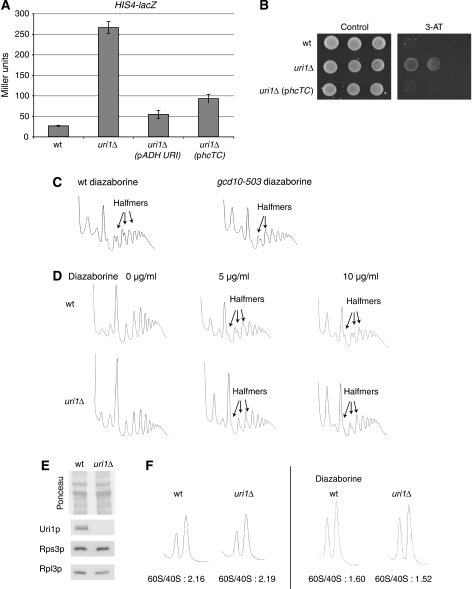
Figure 3.
Uri1p has a function in TC recruitment. (A) Gcn4p activity was determined by measuring β-galactosidase activity of HIS4-lacZ expressed from its own promoter in wild-type (YBL320) or uri1Δ (YBL323) cells transformed with an empty control plasmid (pAD111), or the plasmids pADH-URI (pAD89) or hcTC (pAD116). (B) Serial dilutions of his1-29 gcn2-101 gcn3-101 (YAD37) and his1-29 gcn2-101 gcn3-101 uri1::Nat (YAD379) cells, transformed with an empty vector (pAD111) or hcTC (pAD116) were spotted on plates containing selective medium with or without 5 mM 3-amino triazole (3-AT) to reveal the Gcd– phenotype. Plates were photographed after 4 days at 30°C. (C) gcd10-503 (YAD 36) and isogenic wild-type cells (YAD37) were treated with 10 μg/ml diazaborine for 2 h before incubation with cycloheximide and cell lysis. Cell extracts were loaded on sucrose gradients and polysome profiles were obtained as described for Figure 2D. Halfmer shoulders are indicated by arrows. (D) Wild-type (YAD211) and uri1Δ (YAD219) cells were treated with the indicated amounts of diazaborine for two hours and processed as described for Figure 3C. (E) TCA extracts of wild-type (YAD219) and uri1Δ (YAD211) strains were separated by SDS–PAGE and analysed by ponceau staining (upper panel) or immunoblotting using antibodies specific for Uri1p, Rps3p and Rpl3p. (F) Wild-type (YAD211) and uri1Δ (YAD219) cells were grown without diazaborine (left panel) or with 10 μg/ml diazaborine for 2 h. Cells were lysed without previous cycloheximide treatment in buffer devoid of Mg2+ and extracts were loaded on 7.5–30% sucrose gradients. Polysome profiles were obtained as described earlier, 60S/40S ratios are indicated.
As another indicator of TC levels, we examined diazaborine-induced halfmers in polysome profiles. The drug diazaborine inhibits 27S pre-rRNA processing, leading to a decrease in 60S levels, which in turn produces an excess of 40S subunits and PICs. As a result, cells exposed to diazaborine accumulate additional PICs on actively translating mRNAs, which can be visualized as halfmers (small peaks on the heavy side of the monosomal and polysomal major peaks) in polysome profiles (Pertschy et al, 2004). As PIC formation on mRNAs involves binding of TC to the 40S subunit, we expect that halfmer formation induced by diazaborine should be reduced in mutants harbouring TC-recruitment defects. We tested this assumption using mutants in two well-described initiation factors. Gcd10p is required for tRNAiMet modification, and therefore gcd10-503 cells exhibit reduced levels of TC, leading to derepression of GCN4 (Anderson et al, 1998). Likewise, Gcd6p, a subunit of the initiation factor eIF2B, which acts as a GEF for eIF2 (Bushman et al, 1993), is required for high TC levels. Wild-type cells treated with 10 μg/ml diazaborine induce articulate halfmers, which indeed were reduced in gcd10-503 and to a smaller extent in gcd6-1 cells (Figure 3C; Supplementary Figure 2). Importantly, the same effect could be detected with uri1Δ cells on treatment with 5 or 10 μg/ml diazaborine (Figure 3D). To exclude the possibility that reduced 40S levels may explain this reduced halfmer formation, we first confirmed that the levels of the ribosomal proteins Rpl3p and Rps3p are comparable in uri1Δ compared with wild-type cells (Figure 3E). Moreover, we quantified the 60S/40S ratio in the absence and presence of diazaborine. For this purpose, we performed polysome profiles in the absence of cycloheximide to allow ribosome runoff and in the absence of Mg2+ to dissociate 40S from 60S subunits. As shown in Figure 3F, the 60S/40S ratio of uri1Δ strain is comparable to isogenic wild-type controls, suggesting that the reduction of halfmers on diazaborine treatment is likely caused by reduced PIC levels. Therefore, this result corroborates the interpretation that Uri1p promotes TC binding to 40S subunits in vivo.
To directly test this model, we biochemically measured binding of the TC-component eIF2α to 40S subunits. For this purpose, we treated cells with formaldehyde to cross-link PICs (Valasek et al, 2007), isolated 40S subunits by sucrose gradient centrifugation, and quantified 40S-associated eIF2α levels by western blots using the LI-COR imaging system. Indeed, uri1Δ cells exhibited a subtle but reproducible reduction of eIF2 levels associated with 40S ribosomes compared with wild-type controls (Figure 4A and B). This recruitment defect was further supported by the observation that Rps2p levels in the 40S fractions were significantly higher in uri1Δ cells. A similar effect has been described earlier for eIF1A mutants, which are defective for TC recruitment (Fekete et al, 2005, 2007). Together, these observations suggest that the number of uncharged 40S subunits is higher in uri1Δ cells compared with wild-type controls, supporting the notion that Uri1p promotes TC recruitment to 40S subunits.
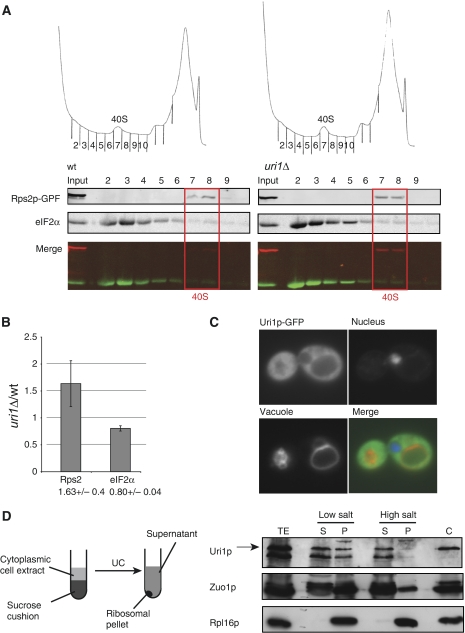
Figure 4.
Uri1p promotes TC recruitment to 40S ribosomal subunits. (A, B) TC levels on 40S subunits were measured by detecting eIF2α in 40S fractions of sucrose gradients. Wild-type (YAD219) and uri1Δ (YAD211) cells were transformed with a plasmid expressing Rps2p-GFP (pAD10). After cross-linking cells with HCHO, extracts were resolved by sedimentation through a 7.5–30% sucrose gradient, and the indicated proteins were detected by immunoblotting and quantified using the LI-COR imaging system. The fractions containing free 40S subunits are boxed. The ‘merge'-panel overlays Rps2p-GFP and the eIF1α signals. (A) Three independent experiments were quantified and shown as average values with standard deviation (B). (C) The subcellular localization of Uri1p–GFP was determined by GFP-fluorescence microscopy in uri1Δ cells (YAD211) transformed with the plasmid pBL22. DNA was visualized by Hoechst-staining, and vacuoles with FM464. (D) Uri1p is loosely associated with ribosomes. Experimental strategy used for ribosomal association assay (left panel). Cell extracts of wild-type cells carrying a plasmid expressing URI1 under control of the ADH-promoter were spun through sucrose cushions containing low (120 mM) or high (800 mM) salt concentrations. Equivalent portions of the supernatant (S), resuspended pellet (P) and total extract (TE) were immunoblotted with the indicated antibodies. A TCA extract prepared from wild-type cells was included as a control (C).
Uri1p associates with ribosomes and interacts with eIF1A (Tif11p)
To examine the subcellular localization of Uri1p, we expressed a fully functional, amino-terminal GFP-fusion protein (GFP–Uri1p) from the GPD-promoter, and analysed its distribution by fluorescence microscopy. As shown in Figure 4C, GFP–Uri1p localized throughout the cytoplasm and was excluded from the nucleus and the vacuole, consistent with a role in protein translation.
We next tested whether Uri1p may associate with cytoplasmic ribosomes. To this end, we separated yeast cytosol into a postribosomal supernatant and a ribosomal pellet in the presence of low or high salt concentrations (Figure 4D). As endogenous Uri1p levels were undetectable in this assay, we overexpressed HA-tagged Uri1p from the constitutive ADH-promoter. Interestingly, a significant fraction of HA-Uri1p was specifically recovered in the ribosomal pellet at low salt concentrations, but released into the supernatant under high salt conditions. As controls, we analysed the ribosome-associated chaperone Zuo1p, which like Uri1p is released under high salt conditions, and the core ribosomal protein Rpl16Ap, which remains stably bound to ribosomal particles under all conditions (Gautschi et al, 2001). These results indicate that at least part of Uri1p associates with ribosomes. The role of Uri1p in TC recruitment could indicate that the ribosome-associated fraction of Uri1p may bind to 40S subunits. However, although Uri1p was present in 40S fractions isolated by sucrose gradients (Supplementary Figure 3), it was not clear whether the presence of Uri1p in these fractions was due to specific association or due to trailing from the upper free protein fractions.
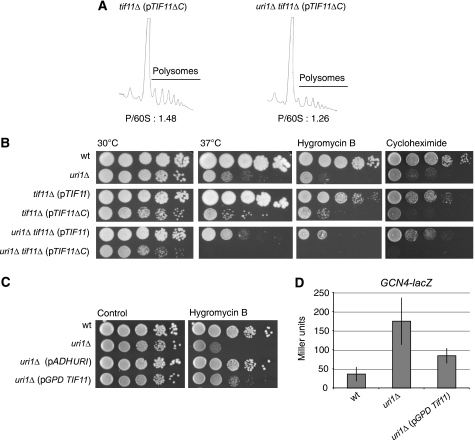
Interestingly, a cytoplasmic yeast two-hybrid screen previously identified a strong interaction between Uri1p and the translation initiation factor eIF1A (encoded by TIF11) (Möckli et al, 2007). eIF1A interacts with the acidic region and PFD domains of Uri1p (Supplementary Figure 4), suggesting that the interaction may be functionally important for translation initiation in vivo. Although TIF11 is an essential gene, deletion of its C terminus confers phenotypes connected to translation initiation and TC recruitment (Fekete et al, 2005), which strongly resemble that of uri1Δ cells. Polysome profiles of uri1Δ tif11ΔC double mutants revealed an additive phenotype compared with each individual mutant (Figure 5A), suggesting that both proteins may cooperate to ensure efficient translation initiation and TC recruitment in vivo. Consistent with this observation, uri1Δ tif11ΔC double mutants were strongly impaired for growth, and hypersensitive to increased temperature stress and exposure to the translation inhibitors hygromycin B and cycloheximide (Figure 5B). Interestingly, we found that overexpression of full-length TIF11 from a GPD-promoter was able to partially suppress the hygromycin B-sensitivity of uri1Δ cells, as shown by serial dilution spottings on rich medium (Figure 5C). Likewise, overexpression of eIF1A was able to suppress expression of the GCN4-lacZ reporter (Figure 5D), suggesting that increased levels of eIF1A can partially bypass the need for Uri1p in translation initiation. Taken together, the functional and physical interaction with eIF1A, which is known to facilitate TC recruitment, support the notion that Uri1p promotes TC binding to 40S subunits.
Figure 5.
Uri1p interacts with eIF1A (Tif11p). (A) Representative polysome traces of tif11Δ (pTIF11ΔC) (YAD407) and uri1Δ tif11Δ (pTIF11ΔC) (YAD399) prepared as described in the legend of Figure 2D. The polysome to 60S ratio was quantified and listed below. (B) Wild-type (YAD211) and uri1Δ (YAD219) cells transformed with an empty vector (pAD48) and tif11Δ (pTIF11) (YAD403), tif11Δ (pTIF11ΔC) (YAD407), uri1Δ tif11Δ (pTIF11) (YAD414) and uri1Δ tif11Δ (pTIF11ΔC) (YAD399) cells were grown in selective medium, and serial dilutions were spotted on YPD plates containing the indicated drugs in the following concentrations: hygromycin B 0.08 mM, cycloheximide 0.07 μg/ml. Plates were grown for 3 days at 30 and 37°C, respectively. (C, D) Wild-type (YAD219) and uri1Δ (YAD211) strains were transformed with an empty vector (pAD48), or plasmids expressing Uri1p from the ADH-promoter (pAD89) or TIF11 from the GPD-promoter (pAD118). Spotting conditions are described in Figure 5B. (D) The strains were also tested for the expression of the GCN4-lacZ reporter construct (pBL199). Error bars represent standard deviations of eight individual transformants.
Uri1p physically interacts with multiple components involved in protein synthesis
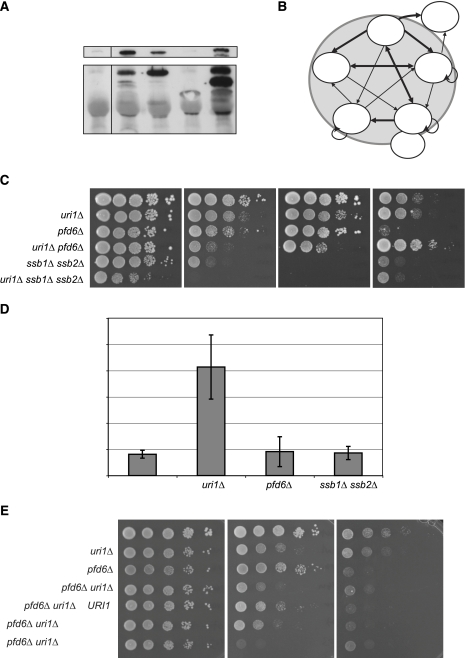
In addition to eIF1A, the cytoplasmic yeast two-hybrid screen identified 14 other Uri1p interactors, among which at least eight can be linked to protein synthesis (Möckli et al, 2007). These candidates include the small ribosomal subunit protein Rps4A, the elongation factor eEF1A, and one of two subunits of eEF1B (Supplementary Table S1). Moreover, several chaperone proteins interacted with Uri1p by two-hybrid assay, including the Hsp70- and Hsp40 family members Ssb1p and Sis1p, both of which have been directly implicated in translation (Nelson et al, 1992; Zhong and Arndt, 1993; Rakwalska and Rospert, 2004). Finally, Uri1p interacted with the peptide-prolyl cis-trans isomerases Fpr1p and Crp1p, which in analogy to the bacterial trigger factor might be involved in folding of newly synthesized proteins. As expected from earlier results (Gstaiger et al, 2003), Uri1p also bound the PFD Pfd6p and the RNA polymerase subunit Rpb5p in this yeast two-hybrid system (Möckli et al, 2007). To biochemically confirm these interactions, we immunoprecipitated the HA-tagged candidate proteins and monitored their binding to myc-tagged Uri1p expressed from its endogenous locus (Möckli et al, 2007). In addition, we co-immunoprecipitated GFP-tagged Uri1p expressed in strains carrying TAP-tagged Ssb1p, Sis1p, eEF1Bγ (Tef4p) and for control Pdf6p, expressed from their endogenous promoters. Indeed, we observed a specific interaction of Uri1p with eEF1A (Tef1p), eEF1Bγ (Tef4p) and Ssb1p (Figure 6A; Möckli et al, 2007).
Figure 6.
Uri1p interacts with proteins involved in protein quality control. (A) Co-immunoprecipitation assays with endogenously tagged Uri1p-interacting proteins. TAP-tagged Pfd6p (YAD349), Ssb1p (YAD155), Sis1p (YAD156), eEF1Bγ (YAD119) and a non-tagged wild-type control strain (YAD219) were transformed with a plasmid expressing URI1-GFP (pBL22) expressed from the GPD-promoter. TAP-tagged proteins were immunoprecipitated, and associated Uri1p–GFP was detected by immunoblotting (upper panel). Arrowheads indicate the immunoprecipitated TAP-tagged proteins; an unspecific band migrating at the same size as Ssb1p-TAP are indicated with an asterisk. (B) Schematic representation of the Uri1p interaction network as revealed by cytoplasmic two-hybrid analysis (for source data see Supplementary Figure 5). Bold arrows mark stronger interactions; arrowheads at both ends indicate that the interaction was detected in both directions. Rounded arrows highlight components that interact with themselves. (C) Wild-type (YAD219), uri1Δ (YAD211), ssb1Δ ssb2Δ (YAD194), pfd6Δ (YAD282), uri1Δ pfd6Δ (YAD262) and ssb1Δ ssb2Δ uri1Δ (YAD234) cells were spotted in serial dilutions on YPD containing as indicated cycloheximide 0.07 μg/ml, hygromycin B 0.08 mM or AZC 1 mg/ml. Plates were incubated at 30 or 37°C for 2 or 3 days. (D) Expression of the GCN4-lacZ reporter construct (pBL199) was measured in wild-type (YAD219), uri1Δ (YAD211), ssb1Δ ssb2Δ (YAD194) and pfd6Δ (YAD282) strains. Error bars represent standard deviations of five individual transformants. (E) Wild-type (YAD219), uri1Δ (YAD211), pfd6Δ (YAD282), uri1Δ pfd6Δ (YAD262) cells carrying an empty vector as well as uri1Δ pfd6Δ cells transformed with plasmids carrying full-length URI1 (pNM100) or the indicated domains (pNM107, pNM103) were spotted in serial dilutions on SD-Leu plates containing as indicated cycloheximide (0.07 μg/ml) or AZC (0.2 mg/ml).
To establish the relationship among the Uri1p-BPs, we examined the interactions among full-length Uri1p, Ssb1p, Sis1p, Pfd6p, Rpb5p and eEF1A by cytoplasmic two-hybrid assays. As negative controls, we used the empty vectors (pASO and NubG), and the three PFDs Pfd4p, Pfd2p and Pfd1p, which did not interact with Uri1p. Interestingly, Ssb1p, eEF1A, Pfd6p and Uri1p formed the core of a small Uri1p interaction network, as each of them showed strong interactions with at least three of the other proteins (Figure 6B; Supplementary Figure 5). Consistent with the co-immunoprecipitation experiments, where the interaction of Uri1p with Sis1p was not detectable, binding of Sis1p to Ssb1p and Pfd6p was stronger compared with its interaction with Uri1p, suggesting that one of these proteins may bridge the interaction of Sis1p and Uri1p. Together, these results indicate that Uri1p functions in a network at the interface of translation and protein quality control together with Ssb1p, eEF1A and Pfd6p.
Functional interaction of Uri1p with the CLIPS Pdf6p and Ssb1/2p
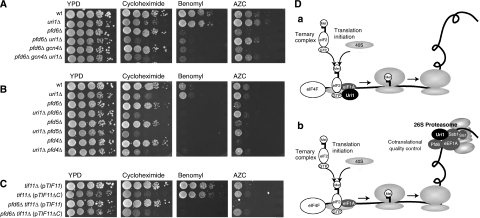
Ssb1/2p and the GimC PFD complex have recently been classified as chaperones linked to protein synthesis (CLIPS) (Albanese et al, 2006). Cells lacking CLIPS function are viable but hypersensitive to translation inhibitors, similar to uri1Δ cells. To examine a possible functional relationship between Uri1p and CLIPS chaperones, we compared the phenotypes of single and uri1Δ double mutants. As expected (Albanese et al, 2006), we found that both the pfd6Δ and ssb1Δ ssb2Δ strains were sensitive to the translation inhibitor cycloheximide and the misfolding-inducing agent AZC (Figure 6C), supporting the notion that they function as CLIPS. The combined deletion strain uri1Δ ssb1Δ ssb2Δ was clearly sicker than the uri1Δ or the ssb1Δ ssb2Δ strains, and this triple deletion strain was extremely sensitive to the translation inhibitor cycloheximide. Similarly, uri1Δ pfd6Δ cells were hypersensitive to cycloheximide, suggesting that the functions of Uri1p and these chaperones in protein translation are additive. In accordance with this result, the chaperone mutants did not affect GCN4-lacZ expression (Figure 6D), indicating that unlike Uri1p they are not involved in TC recruitment. Moreover, in contrast to uri1Δ, strains carrying deletions in any of the PFD genes are sensitive to the microtubule-depolymerizing drug benomyl (Figure 7B), consistent with the known role of the GimC PFD complex in tubulin folding (Geissler et al, 1998). Although ssb1Δ ssb2Δ and pfd6Δ cells were sensitive to AZC, uri1Δ ssb1Δ ssb2Δ cells were slightly resistant to AZC compared to the ssb1Δ ssb2Δ strain. Strikingly deletion of URI1 restored full resistance of pfd6Δ cells to this drug. In contrast to the PFD domain, this effect was reverted by plasmids expressing full-length or the acidic domain of Uri1p (Figure 6E). Importantly, this AZC resistance is not altered by deletion of GCN4 (Figure 7A), excluding the possibility that Gcn4p-induced increase of prolin biosynthesis (Li and Brandriss, 1992) might reduce incorporation of the prolin analog AZC. To test whether suppression of AZC sensitivity by deletion of URI1 was specific for Uri1p interactors, we compared AZC sensitivity of strains deleted for other PFDs that do not physically interact with Uri1p (Supplementary Figure 6), either singly or in combination with deletion of URI1. Similar to pfd6Δ, deletion of URI1 also suppressed the AZC sensitivities of pfd4Δ and pfd5Δ cells (Figure 7B), suggesting that this phenotype may be explained by a general mechanism such as the reduction of translation initiation. Indeed, the tif11ΔC mutation, which causes a strong translation initiation defect, also suppressed AZC sensitivity of pfd6Δ cells (Figure 7C).
Figure 7.
Suppression of AZC sensitivity of pfd6Δ cells by deletion of URI1 may be caused by a slowdown in translation. (A) Wild-type (YAD219), uri1Δ (YAD211), pfd6Δ (YAD282), uri1Δ pfd6Δ (YAD262), pfd6Δ gcn4Δ (YAD485) and pfd6Δ gcn4Δ uri1Δ (YAD460) strains were spotted in serial dilutions on YPD plates containing the indicated drugs AZC (1 mg/ml), cycloheximide (0.07 μg/ml), benomyl (25 μg/ml) and incubated for 3 days at 30 or 37°C, respectively. (B) Serial dilutions of wild-type (YAD219), uri1Δ (YAD211), pfd6Δ (YAD282), uri1Δ pfd6Δ (YAD262), pfd4Δ (YAD426), uri1Δ pfd4Δ (YAD428), pfd5Δ (YAD291) and uri1Δ pfd5Δ (YAD431) strains were spotted on YPD plates as described for panel (A). (C) tif11Δ (pTIF11) (YAD462), tif11Δ(pTIF11ΔC) (YAD464), pfd6Δ tif11Δ(pTIF11) (YAD468), pfd6Δ tif11Δ(pTIF11ΔC) (YAD470) strains were spotted under the same conditions as described in (A) and (B). (D) A model illustrating the functions of Uri1p in protein translation. (a) Uri1p promotes TC loading during translation initiation and interacts with the translation initiation factor eIF1A. (b) In addition, Uri1p interacts with several components involved in cotranslational quality control. We speculate that misfolded nascent chains may recruit Uri1p to the cotranslational quality control machinery, which in turn might lead to reduction of the translation initiation function of Uri1p. Black, Uri1p; dark grey, Uri1p interactors; white, other factors involved in translation initiation.
Discussion
In this study, we show that yeast Uri1p is required for efficient translation initiation, most likely by promoting TC recruitment to 40S subunits. Moreover, Uri1p interacts with eEF1A and the CLIPS Ssb1,2p and Pfd6p, which are involved in folding and degrading newly synthesized proteins. Our results thus suggest that Uri1p may mediate between translation initiation and quality control.
Uri1p regulates translation initiation by promoting recruitment of TC to 40S subunits
Translation initiation requires that the TC, which consists of the methionine-charged initiator tRNA and GTP-bound initiation factor eIF2, is recruited to the 40S subunit. Several lines of evidence strongly suggest that Uri1p promotes this step. First, GCN4 translation is upregulated in uri1Δ cells under rich conditions, and this phenotype is suppressed by a plasmid overexpressing all components of the TC (Figure 3A and B). Second, the formation of aberrant halfmers induced by diazaborine treatment is reduced in uri1Δ cells (Figure 3D). Halfmers in polysome profiles reflect mRNAs with one or several ribosomes and an additional 40S subunit and can be caused by a reduction of 60S subunits or an excess of 40S subunits. As the ratio of the 60S and 40S peaks in polysome profiles, and Rpl3p and Rps3p levels by western blots are unchanged in wild-type and uri1Δ cells (Figure 3E and F), the first two possibilities are unlikely. Moreover, because the free 40S peak on diazaborine treatment is increased in uri1Δ compared with wild-type cells (Figure 3D), a negative effect on 40S ribosome biogenesis is unlikely. However, the reduction of halfmers on diazaborine treatment can be explained by inefficient TC recruitment to 40S subunits, which leads to reduced PIC levels, thus supporting a role of Uri1p in TC loading. Third, sucrose-gradient analysis of cross-linked protein extracts revealed reduced levels of the TC component eIF2α in 40S fractions in uri1Δ cells (Figure 4A and B). This relatively modest biochemical effect indicates that Uri1p is not a central translation initiation factor, but rather increases the efficiency of this process. Finally, Uri1p genetically and physically interacts with the translation initiation factor eIF1A (Figure 5), which was earlier found to function in TC recruitment, PIC assembly, ribosomal scanning and AUG selection (Olsen et al, 2003; Fekete et al, 2005, 2007). The carboxy-terminus of eIF1A is required for efficient TC recruitment (Olsen et al, 2003; Fekete et al, 2005), and this defect is worsened in cells lacking Uri1p function. Conversely, overexpression of eIF1A can partially suppress the hygromycin B sensitivity of uri1Δ cells (Figure 5C), suggesting that Uri1p and eIF1A have overlapping functions in the recruitment of TC to the 40S subunit (PIC assembly). Taken together, our results thus suggest that Uri1p may support translation initiation by directly promoting TC recruitment and/or activating initiation factors such as eIF1A. Although under normal conditions lack of Uri1p results in a global reduction of translation initiation, Uri1p is required to cope with stress conditions such as high temperature or the presence of translation inhibitors (Figure 2B).
Uri1p interacts with CLIPS
Our results together with earlier published data (Gstaiger et al, 2003; Möckli et al, 2007) also revealed a physical and functional interaction of Uri1p with several factors involved in protein synthesis downstream of translation initiation, including the translation elongation factors eEF1A (Tef1p, Tef2p) and eEF1Bγ (Tef4p), the Hsp70 chaperone Ssb1, 2p and the Gim-complex component Pfd6p. Both Pfd6p and Ssb1, 2p belong to a set of chaperones called CLIPS, which in contrast to heat-shock chaperones, but similar to various translation components, are downregulated at the transcriptional level in response to heat shock (Albanese et al, 2006). Interestingly, URI1 is also downregulated on heat-shock treatment as judged by microarray data (Gasch et al, 2000). However, in contrast to cells deleted for bona fide members of the CLIPS family, uri1Δ cells are not sensitive to the misfolding-inducing agent AZC (Figures 6 and 7). Moreover, unlike deletion strains of all yeast PFDs, uri1Δ cells are not sensitive to the presence of the microtubule-destabilizing drug benomyl (Figure 7A–C). Finally, the PFD domain is not required for the function of Uri1p in vivo (Figure 2C). Together, these data suggest that Uri1p may not directly participate in protein folding, but rather functions as a regulator at the interface between translation and protein quality control.
Uri1p may link translation initiation with cotranslational quality control
What could be the role of a non-essential factor promoting translation initiation that interacts with several components of the quality control machinery? It is possible that Uri1p is involved in a feedback mechanism that actively prevents translation initiation under conditions when misfolded proteins accumulate. For example, Uri1p may be recruited to nascent chains by its interactors eEF1A (Tef1p), Pfd6p, Ssb1,2p and Sis1p under conditions when increased levels of misfolded proteins are being synthesized, thereby reducing translation initiation (Figure 7D). Indeed, reducing translation initiation by deletion of URI1 or eIF1A mutations restores growth of chaperone mutants such as pdf6Δ cells on media containing AZC. A comparable feedback mechanism known as the ‘unfolded protein response' (UPR) exists in the endoplasmatic reticulum of higher eukaryotes. Accumulation of unfolded proteins in the ER dissociates the Hsp70 chaperone BIP from the transmembrane kinase PERK, which in turn phosphorylates and inhibits eIF2α in the cytosol and thereby inhibits translation initiation by the same mechanism as scGcn2p (Harding et al, 2000; reviewed in Schroder and Kaufman, 2005).
In addition to promoting correct folding of newly synthesized proteins, quality control mechanisms also trigger degradation of misfolded proteins. In the endoplasmatic reticulum, the UPR is closely coupled to ERAD (ER-associated degradation) (reviewed in Buck et al, 2007). Likewise, nascent peptides in the cytoplasm can be ubiquitinated cotranslationally (references in Chuang and Madura, 2005), but it remains unclear how the ubiquitination machinery recognizes these proteins. A role in cotranslational protein degradation has been suggested earlier for the Uri1p-interacting proteins eEF1A (Tef1p, Tef2p) as well as Ssb1,2p and Sis1p (Zhong and Arndt, 1993; Gonen et al, 1994; Ohba, 1997; Rakwalska and Rospert, 2004; Chuang et al, 2005). It is interesting to note in this context that human Uri1p was identified as a protein interacting through STAP1 with Skp2p, which functions in a cullin1-based E3 ubiquitin ligase (Gstaiger et al, 2003). It is thus possible that Uri1p may help to recruit the ubiquitination machinery to mistranslated proteins by interacting with Ssb1,2p, eEF1A and Pfd6p. This might lead to reduced levels of Uri1p functioning in translation initiation and contribute to slowing down translation initiation to allow cells to cope with difficult conditions.
Materials and methods
Strains (Supplementary Table S2) and plasmids (Supplementary Table S3) are described in Supplementary data.
Antibodies
The following antibodies were used: Actin (Chemicon), myc (Gramsch Laboratories), PAP (peroxidase anti-peroxidase) (Sigma), GFP (Roche), HA-11 (Convance), eIF2α-P (Genetex), Zuo1p (S Rospert), Rpl16p (S Rospert), Sui2p (T Dever), Rps3 (M Seedorf), Rpl3 (J Warner). Uri1p-specific antibodies were generated by immunizing rabbits with a recombinant Uri1p-fragment encompassing amino acids 165–331. Antibodies were affinity purified and characterized as shown in Figures 1D and 3E.
LacZ-assays
β-galactosidase activity in strains bearing a GCN4-lacZ (uORFs 1–4) (pBL199) or xxxxGCN4-lacZ (mutated uORFs) (pBL201) reporter plasmid or strains carrying endogenous HIS4-lacZ (YBL320) was measured using standard assays detecting ONPG (o-nitrophenyl-β-D-galactoside) cleavage by β-galactosidase, as described earlier (Valtz and Peter, 1997). β-galactose levels were quantified as Miller units (OD600 × 1000)/(OD420 × min), where OD600 indicates the density of the culture and absorption at OD420 denotes the amount of cleaved ONPG in a given time [min].
Polysome analysis and diazaborine experiments
Yeast cultures (100 or 200 ml) were grown to exponential phase, and treated with 100 μg/ml cycloheximide for 10 min on ice before centrifugation. Pellets were washed twice and resuspended in 500–800 μl lysis buffer (10 mM Tris–HCl (pH 7.5), 100 mM NaCl, 30 mM MgCl2, 100 μg/ml cycloheximide, 0.2 mg/ml heparin, 0.2 μl/ml DEPC). Cells were lysed by bead-beating and lysates (10 OD260 absorption units) were loaded on 12.2 ml 15–50% linear sucrose gradients prepared in 50 mM Tris–acetate (pH 7.5), 50 mM NH4Cl, 12 mM MgCl2). The gradients were centrifuged at 39 000 rpm in a Beckman SW41 rotor at 4°C for 2 h 45 min, followed by analysis with an ISCO UV-6 gradient collector. The polysome to 80S ratio (P/80) was determined using ImageJ software (http://rsb.info.nih.gov/ij/).
For diazaborine experiments, 50 ml cultures were grown to OD600 0.5, at which time 5 or 10 μg/ml diazaborine (kindly provided by Gregor Hoegenauer) was added for 2 h at 30°C. Cycloheximide was added to a final concentration of 100 μg/ml, and polysome profiles were obtained as described above.
To determine the 60S/40S ratio by polysome profiling, 100 ml wild-type (YAD219) and uri1Δ (YAD211) cells were grown to exponential phase and incubated with 10 μg/ml diazaborine without cycloheximide addition before lysis. Cells were lysed using the same procedure as described above, but MgCl2 was omitted in the lysis buffer. Extracts were loaded on a 7.5–30% sucrose gradient, centrifuged and analysed as described.
PIC cross-linking using formaldehyde (HCHO)
In total, 100 ml wild-type (YAD219) and uri1Δ (YAD211) cells transformed with a plasmid expressing Rps2p-GFP (pAD10) were grown to exponential phase and incubated on ice with 1% formaldehyde for 1 h as described (Valasek et al, 2007). Cross-linking was stopped by the addition of glycine to 0.1 M final concentration. Cells were centrifuged, and one volume cell was resuspended in one volume lysis buffer (20 mM Tris (pH 7.5), 50 mM KCl, 10 mM MgCl2, protease inhibitor cocktail (Roche), 5 mM NaF, 1 mM DTT, 1 mM PMSF). Subsequently, cells were lysed with one volume glass beads using a beat beater. About 20 OD260 units of lysate were applied on 7.5–30% sucrose-gradients and centrifuged for 5 h at 41 000 rpm. A measure of 500 μl fractions were collected and precipitated by addition of trichloracetic acid (TCA) to a final concentration of 20%. Pellets were washed in ice-cold acetone and resuspended in loading buffer. Western blot signals were quantified using the LI-COR imaging system and ImageJ software. Input-ratios (Rps2input(wt)/Rps2input(uri1Δ) and eIF2αinput(wt)/eIF2αinput(uri1Δ)) were used to normalize the 40S signals and adjust differences in loading. Then, ratios of 40S fractions were calculated (Rps240S(uri1Δ)/Rps240S (wt) and (eIF2α40S(uri1Δ)/eIF2α40S(wt)). Average values and standard deviation of three independent experiments were calculated.
Microscopy
Localization of pGPD-GFP-URI1 was determined in uri1Δ cells, which were grown to exponential phase on minimal medium lacking uracil at 30°C. Vacuoles were stained with FM460 (final conc. 16 μM), incubated for 30 min at 30°C, followed by washing with SD-Ura medium and further incubation for 30 min at 30°C. During the second incubation, the DNA-stain Hoechst (Hoechst 33342) was added (2 μg/ml final conc.). Samples were analysed on a Zeiss Axiovert 200 M fluorescence microscope.
Ribosome-binding assay
Ribosome binding of Uri1p was examined by ultracentrifugation through a sucrose cushion containing low (120 mM) or high (800 mM) concentrations of potassium acetate. Wild-type (YAD219) and uri1Δ (YAD211) cells were grown in 100 ml selective medium, pelleted and resuspended in 2 ml lysis buffer (10 mM Tris pH 7.5, 50 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% TritonX-100, 1 mM DTT, protease inhibitor cocktail (Roche)). Cells were broken using a one shot cell disrupter (constant systems LTD), and the lysate was cleared by centrifugation at 4°C for 15 min at full speed in an eppendorf-centrifuge (5417R), followed by 30 min spinning at 80 000g using a TLA 120.2 rotor in a Beckman coulter Optima LE-80K ultracentrifuge. A measure of 60 μl extract were removed for the ‘total'-sample, and 60 μl were loaded on a 90 μl low-salt (120 mM potassium acetate) or high-salt (800 mM potassium acetate) sucrose cushion (25% sucrose, 2 mM DTT, 5 mM magnesium acetate, protease inhibitor cocktail (Roche)). After centrifugation for 2–4 h at 200 000g in a TLA100 rotor, cytoplasmic supernatants were collected and ribosomal pellets resuspended in 1 × buffer (2 mM DTT, 5 mM magnesium acetate, 120 mM potassium acetate). ‘Total'-samples, supernatant fractions and resuspended pellets were precipitated by TCA (final concentration 5%) and resuspended in loading buffer. Corresponding amounts were analysed by SDS–PAGE and immunoblotting.
Immunoprecipitation experiments
For immunoprecipitation of endogenously TAP-tagged proteins, 100 ml yeast cultures were grown and one cell volume was resuspended in one volume RIPA buffer (5 mM HEPES pH 7.4, 1% Triton, 1% NaDeoxycholate, 0.1% SDS, 100 mM NaCl, protease inhibitor cocktail (Roche), 1.8 mM NaVa, 72 mM β-glycerophosphate) and lysed by bead beating. Equivalent amounts of protein (around 2–3 mg in 300 μl) was added to 30 μl IgG Sepharose beads (GE Healthcare) and incubated at 4°C for 90 min. Beads were washed five times with RIPA buffer before elution in 40 μl loading buffer.
Supplementary Material
Supplementary Information
Acknowledgments
We thank G Hoegenauer for the diazaborine, A Hinnebusch, K Tatchell for providing plasmids and strains, and S Rosbert, T Dever, M Seedorf and J Warner for antibodies. We are grateful to C Rupp for technical support, members of the Peter group and P Linder for stimulating discussions, and R Dechant and I Zemp for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation (SNF), the Competence Center for Systems Physiology and Metabolic Diseases (SPMD) and the ETH Zurich.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG (1998) The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 18: 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Wright CM, Brodsky JL (2007) The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev Biol 18: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman JL, Asuru AI, Matts RL, Hinnebusch AG (1993) Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol 13: 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K (2005) Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SM, Madura K (2005) Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics 171: 1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Hinnebusch AG, Tamame M (1998) Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 148: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE (2002) Gene-specific regulation by general translation factors. Cell 108: 545–556 [DOI] [PubMed] [Google Scholar]
- Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W (2007) S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell 28: 28–40 [DOI] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG (2005) The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J 24: 3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG (2007) N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J 26: 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S (2001) RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci USA 98: 3762–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E (1998) A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17: 952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, Chakraburtty K, Schwartz AL, Ciechanover A (1994) Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci USA 91: 7648–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W (2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science (New York, NY) 302: 1208–1212 [DOI] [PubMed] [Google Scholar]
- Harashima S, Hinnebusch AG (1986) Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 6: 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Li W, Brandriss MC (1992) Proline biosynthesis in Saccharomyces cerevisiae: molecular analysis of the PRO1 gene, which encodes gamma-glutamyl kinase. J Bacteriol 174: 4148–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckli N, Deplazes A, Hassa PO, Zhang Z, Peter M, Hottiger MO, Stagliar I, Auerbach D (2007) A yeast split-ubiquitin based cytosolic screening system (cytoY2H) to detect interactions between transcriptionally active proteins. Biotechniques 42: 725–730 [DOI] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45: 201–207 [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA (1992) The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105 [DOI] [PubMed] [Google Scholar]
- Ohba M (1997) Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett 409: 307–311 [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG (2003) Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Zisser G, Schein H, Koffel R, Rauch G, Grillitsch K, Morgenstern C, Durchschlag M, Hogenauer G, Bergler H (2004) Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol Cell Biol 24: 6476–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwalska M, Rospert S (2004) The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol Cell Biol 24: 9186–9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789 [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404: 770–774 [DOI] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU (1999) Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J 18: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Varshavsky A (2000) Detecting and measuring cotranslational protein degradation in vivo. Science (New York, NY) 289: 2117–2120 [DOI] [PubMed] [Google Scholar]
- Valasek L, Szamecz B, Hinnebusch AG, Nielsen KH (2007) In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol 429: 163–183 [DOI] [PubMed] [Google Scholar]
- Valtz N, Peter M (1997) Functional analysis of FAR1 in yeast. Methods Enzymol 283: 350–365 [DOI] [PubMed] [Google Scholar]
- Zhong T, Arndt KT (1993) The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell 73: 1175–1186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information