Figure 4. The βTRCP-REST pathway controls neural differentiation.
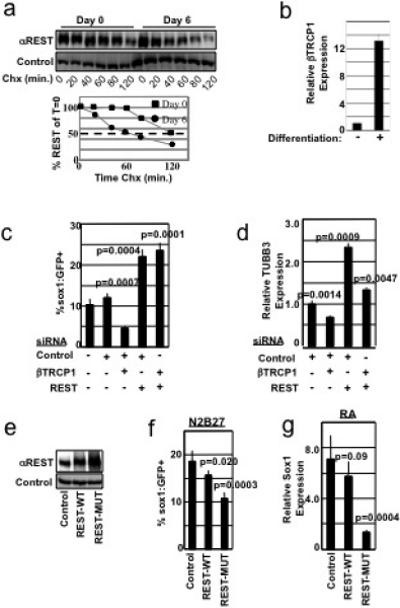
a, ES cells differentiated for 0 or 6 days were examined for REST protein half-life in a cycloheximide (Chx) timecourse. Differentiated lysates were analyzed at 3x the concentration of undifferentiated lysates. Quantitation of REST levels is shown in the lower panel. b, ES cells were differentiated for 0 or 6 days. βTRCP1 mRNA was analyzed by qRT-PCR, and normalized to GAPDH mRNA abundance. Experiment was performed in triplicate (error bars +/− s.d.). c, 46C cells transfected with the indicated combination of siRNAs were differentiated in N2B27 medium and analyzed for Sox1:GFP expression by flow cytometry. Experiments were performed in quadruplicate (error bars +/− s.d.) and is representative of 4 independent experiments. d, 46C cells from c were analyzed for expression of TUBB3 mRNA by qRT-PCR, and normalized to GAPDH mRNA abundance. Experiments were performed in triplicate (error bars +/− s.d.). e, 46C cells expressing control, Flag-REST-WT, or Flag-REST-MUT (triple point mutation in the REST-degron) cDNA were immunoblotted with αREST (upper panel) or αvinculin (lower panel) antibodies. Note: In this experiment, exogenous REST was expressed at levels higher than endogenous REST. f, 46C cells from e were cultured in N2B27 differentiation medium and analyzed for Sox1:GFP fluorescence. This experiment was performed in sextuplicate (error bars +/− s.d.) and is representative of 2 independent experiments. g, ES cells were infected as in e, differentiated into the neural lineage using an embryoid body-retinoic acid protocol, and analyzed for Sox1 mRNA by qRT-PCR (normalized to GAPDH mRNA abundance). Experiment was performed in triplicate (error bars +/− s.d.).
