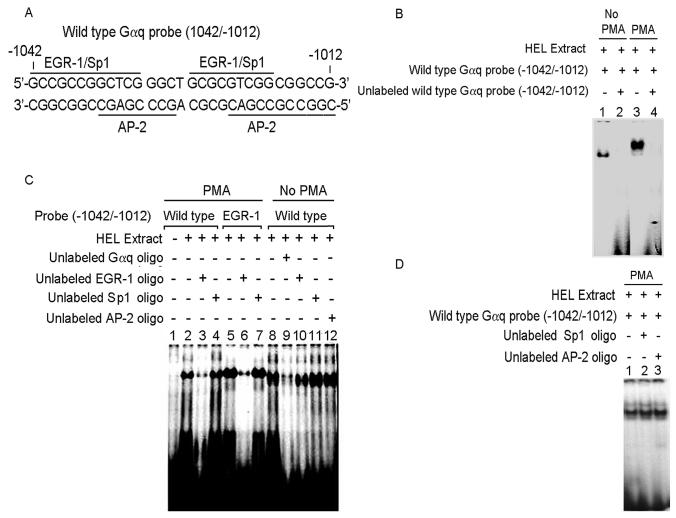
Figure 2. Electrophoretic mobility shift assay (EMSA) with Gαq oligonucleotide (−1042/−1012) and nuclear proteins from untreated and PMA-treated HEL cells.
(A) Gαq oligonucleotide sequence showing two predicted EGR-1 sites overlapping with Sp1 and AP-2 sites. (B) EMSA with Gαq oligonucleotide and nuclear extracts from untreated (Lanes 1-2) and PMA-treated (Lanes 3-4) HEL cells. Lanes 1 and 3: protein complex formed between the Gαq oligonucleotide and HEL nuclear extracts. Lanes 2 and 4: competition with excess unlabeled Gαq oligonucleotide indictating the specificity of binding. (C) EMSA with Gαq oligonucleotide with PMA-treated (Lanes 1-7) and untreated (Lanes 8-12) nuclear extracts and competition with EGR-1, Sp1 and AP-2 consensus oligos. Lane 1: labeled oligonucleotide probe alone. Lane 2: binding with PMA-treated nuclear extract. Lane 3: competition with excess unlabeled EGR-1 oligo. Lane 4: no competition with excess unlabeled Sp1 oligo. Lane 5: protein complex formed between labeled EGR-1 oligo and PMA-treated extract. Lane 6: competition with excess unlabeled EGR-1 oligo. Lane 7: no competion with Sp1 oligo. Lane 8: binding with untreated nuclear extract. Lane 9: competition with excess unlabeled oligonucleotide. Lanes 10-12: no competition with EGR-1, Sp1 and AP-2 consensus oligos respectively. (D) Lane 1: binding between Gαq oligonucleotide and PMA-treated nuclear extract. Lanes 2 and 3: no competition with Sp1 and AP-2 consensus oligos.