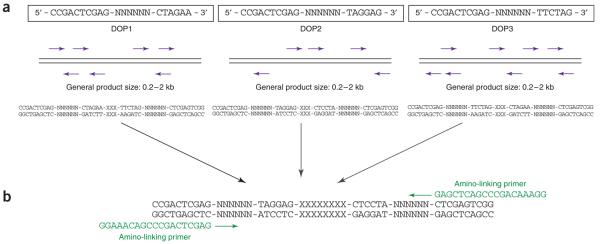
Figure 2.
Schematic overview of target DNA preparation for array production (a) DOP-PCR: The variation of six consecutive bases within the primer (N) and the low annealing temperature during the first amplification cycles allow the primers to bind along the template DNA. For each primer (DOP1, DOP2 and DOP3), the amplification efficiency relies on the frequency with which bases in the target sequence match the six 3′ end bases of the primer. (b) Amino-linking PCR: Amplification of the target DNA by DOP-PCR is followed by a secondary PCR using a 5′ amine-modified primer. The 3′ end of the amino-linked primer matches the 5′ end of each of the three DOP-PCR primers (here only the amplification of DOP2-derived PCR products is shown as an example).