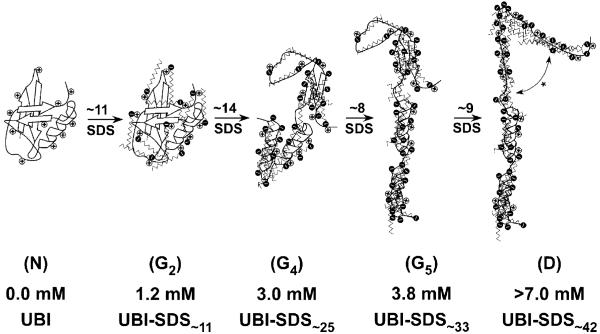
Figure 7.
Pictures describing the hypothetical structure of UBI-SDSn complexes within groups N, G2,G4,G5, and D. The 11 cationic sites (e.g., 7 lysines and 4 arginines; we consider the unique histidine and the amino N-terminus to be uncharged at pH 8.4) are represented by positive charges along the protein backbone. N represents the native UBI (PDB 1UBQ; one turn of the α-helix contains four amino acids). G2 is a native-like UBI-SDS complex (as revealed by CD, Figures 2a,b and S4a) associated with 11 bound SDS molecules (as determined from the charge ladder analysis, Figure 4e). G4 is less organized (perhaps a molten globule, Figure 2c), obtained after the binding of ∼14 SDS to G2 (cf. Figure 6b) with the conversion of about half of the β-stands into α-helices (cf. Figure 2b). Transitions from G4 to G5 and from G5 to D respectively involve ∼8 and ∼9 molecules of SDS (cf. Figures 5 and 6b) with no major change in secondary structure as revealed by CD; however, D might be best represented by an unfolded, SDS-saturated polypeptide with extensive α-helical structure, represented in this cartoon by the disruption of the remaining β-strand in G5 (*).