Short abstract
Next-generation sequencing technology can be used to assess rates and regulation of translation across the entire yeast transcriptome.
Abstract
Next-generation massively parallel sequencing technology provides a powerful new means of assessing rates and regulation of translation across an entire transcriptome.
The introduction of massively parallel DNA sequencing platforms over the past five years - so-called 'next-generation' sequencing technology - has created the capacity to generate tens of millions of short sequence reads in a single run. These sequences can be identified by alignment to the known genomes of the all-important model organisms, including Homo sapiens [1]. The information garnered from this technology is providing new insights into important areas of genome, chromatin and transcriptome biology.
One of the applications of next-generation sequencing - short-read cDNA analysis or 'RNAseq' [2,3] - has its conceptual roots in serial analysis of gene expression (SAGE) [4]. Whereas SAGE provides thousands of sequences of short sequence tags that have been cloned as concatemers, RNAseq ups the ante to tens of millions of independently derived sequences per experiment. For RNA biology, transcriptome analysis by RNAseq provides robust quantitative reproducibility, dynamic range of many orders-of-magnitude, transcript directionality, analysis of repetitive sequences, independent measurement of highly similar sequences and detection of post-transcriptional processing at the single nucleotide level. Using RNAseq methodology, a recent study from Jonathan Weissman's laboratory (Ingolia et al. [5]) yielded a snapshot of the steady-state linear distribution of ribosomes on RNA transcripts in cells of Saccharomyces cerevisiae, providing a new powerful experimental tool for analysis of translational control and co-translational processes.
Ribosome profiling by RNAseq
Following the early demonstration by Steitz of ribosome footprints at the initiation codons of bacteriophage R17 RNA [6], Wolin and Walter showed that eukaryotic ribosomes carrying out translation protected around 30 nucleotides of mRNA sequence from digestion by RNase [7]. Exploiting this observation, they demonstrated clusters of ribosome protection at discrete sites in the preprolactin transcript. These clusters were interpreted as reflecting rate-limiting steps at translation initiation and termination, as well as ribosome pausing at the site of interaction of the nascent signal peptide with the signal recognition particle.
Ingolia et al. [5] have now extended analysis of these ribosome-protected fragments to the genome-wide scale through RNAseq technology. They implemented an imaginative intramolecular ligation strategy to generate directional, unbiased cDNA libraries for sequencing ribosome-protected RNA fragments. Despite significant contamination by ribosomal RNA, they were able to assign 7 × 106 RNAseq reads to more than 4,500 yeast genes. These ribosome 'footprints' were mapped with a high degree of precision and revealed a remarkable three-base periodicity corresponding to the codons within protein-coding sequences across the transcriptome. The abundance of ribosome-protected fragments from a given gene was used to predict the level of the encoded protein and was shown to be a significantly better predictor than mRNA level (multiple regression correlation coefficient R2 = 0.42 versus R2 = 0.17).
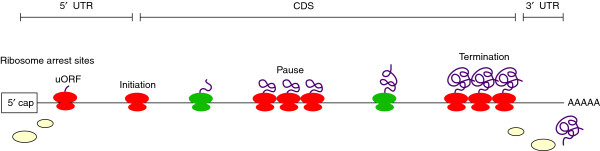
This study also demonstrated how patterns of ribosome footprints could be used to provide insights into translational regulatory mechanisms. Figure 1 illustrates potential sites of ribosome localization on a generic mRNA. From the Wolin and Walter study [7], one would anticipate footprints at initiation codons and perhaps enhanced ribosome density at termination sites.
Figure 1.
Positioning of ribosomes on a messenger RNA. The 5' cap is to the left and the poly(A) tail is to the right. The red symbols depict non-random accumulation of ribosomes at an uORF, the initiation codon, a site of ribosome pausing within the coding sequence (CDS), and at termination. The green symbols represent freely translating ribosomes at random sites along the coding sequence.
Ribosomes would be expected to distribute randomly across coding sequences, with the exception of the codon periodicity noted above. Non-random occurrences of footprints within coding sequences are interpreted as sites of translational pausing, for example those associated with rare codons or co-translational activities. Within the untranslated terminal regions (UTRs) of mRNA, footprints might be expected in association with functional upstream open reading frames (uORFs). Indeed, as expected, Ingolia et al. [5] find that 98.8% of the footprints mapped to coding sequences, with the remainder predominantly associated with uORFs in the 5' UTRs.
Although uORFs are known to participate in translational control [8], the extent of their translation across a transcriptome has never been evaluated. To attempt this, Ingolia et al. [5] annotated a total of 1,048 candidate uORFs with AUG starts in the yeast transcriptome and found that 153 of these showed evidence of ribosome association under the growth conditions examined. Among these ribosome-associated uORFs was the gene GCN4. Ribosome footprints over the four uORFs in GCN4 behaved upon amino acid starvation as predicted by the generally accepted model [9] for regulation of this gene - uORF 1 is constitutively translated and there is a reciprocal relationship between translation of uORFs 2-4 and the main coding sequence that is controlled by amino acid starvation.
Interestingly, regulated ribosome loading, apparently originating from two non-AUG starts, was observed upstream of the known uORFs in the GCN4 5' UTR. Although the existence of uORFs with non-AUG initiation codons has been the subject of speculation, the presence of these in GCN4, as well as in more than 1,600 other candidates highlighted by Ingolia et al. [5], gives fascinating hints of previously unrecognized modes of translational control.
Perspectives and cautions
Ribosome profiling by RNAseq is certain to uncover many new and unexpected aspects of mRNA translation and its regulation. The most straightforward application will result from more robust prediction of protein levels than can be obtained from transcript abundance alone [5]. Even more significant will be new insights into the events that occur as a ribosome traverses an mRNA from the cap to the poly(A) tail. A striking example of this in the work of Ingolia et al. is the apparent abundance of uORFs with non-AUG starts throughout the yeast transcriptome. The implications of these new insights for both translational control and constitutive translation efficiency are tremendous. New clues regarding the events that occur as ribosomes pause along the coding sequences are likely to emerge after more extensive analysis of the existing data and/or increasing the sequence depth. Such co-translational processes might include folding or insertion of nascent peptides into cellular structures, as well as non-standard decoding mechanisms such as frameshifting or readthrough of termination codons.
As with any powerful new methodology, the results should be interpreted with caution; there are undoubtedly pitfalls awaiting the unwary. For example, one should be prepared for regulated changes in 5' UTR structure, which may occur commonly in yeast [10,11] and perhaps other species. These changes in UTR structure could drastically alter patterns of ribosome footprints. Likewise, the mere presence of a ribosome on a coding sequence does not mean that it is elongating its nascent polypeptide chain. A polyribosome with all ribosomes arrested at random would show footprints indistinguishable from those of an actively translating polysome. Regulation at the level of elongation is particularly relevant in the context of current controversy over the mechanisms by which microRNAs inhibit translation [12-15].
A technical issue could also drastically influence the interpretation of results. Before preparing extracts, it is routine procedure in many labs to 'freeze' the ribosomes on transcripts with high concentrations of the elongation inhibitor cycloheximide. If the concentration of the inhibitor is not sufficient, elongation is preferentially inhibited over initiation (at least in mammalian cells) and ribosomes are loaded onto transcripts [16], an artifact that the resolving power of RNAseq profiling would easily detect. Considering that ribosomes 'read' mRNA at a rate of about ten codons per second [17], exposure to intermediate concentrations of cycloheximide for only a few seconds (as a result of inefficient uptake or delivery of the inhibitor), would severely distort the distribution of ribosomes on transcripts, resulting in a higher density at the 5' end of the coding sequence. This technical problem should be particularly noted in experiments with intact animals, where delivery of the inhibitor is less controllable. The foregoing are simply words of caution, however, and should not detract from the power and elegance of this new experimental approach.
When it comes to defining mechanisms of translational control, the results of ribosome profiling by RNAseq complement the information obtained by analysis of polyribosomes using techniques involving physical separation. A simple example illustrates this point. If the ribosome "density" (as defined by Ingolia et al. [5]) is found to decrease by a factor of ten for a particular transcript, two interpretations come to mind: all of the transcripts are being translated at 10% the rate (that is, the rate of initiation has dropped by 90%); or 10% of the transcripts are being translated with the remainder in untranslated messenger ribonucleoprotein particles. RNAseq profiling does not distinguish between these alternatives. With currently available technologies, precise mechanisms of translational control can only be defined by combining the extraordinary power of RNAseq profiling with the kinds of information obtained from traditional polysome profiles generated by sucrose gradient centrifugation or other physical separation methods.
Acknowledgments
Acknowledgements
I would like to thank Alan Weiner, Adam Geballe and Vivian MacKay for critically reading the manuscript and providing insightful suggestions.
References
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Grimmond SM. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. doi: 10.1186/gb-2008-9-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA. Discriminatory ribosome rebinding of isolated regions of protein synthesis initiation from the ribonucleic acid of bacteriophage R17. Proc Natl Acad Sci USA. 1973;70:2605–2609. doi: 10.1073/pnas.70.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/MCB.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Law GL, Bickel KS, MacKay VL, Morris DR. The under-translated transcriptome reveals widespread translational silencing by alternative 5' transcript leaders. Genome Biol. 2005;6:R111. doi: 10.1186/gb-2005-6-13-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel KS, Morris DR. Role of the transcription activator Ste12p as a repressor of PRY3 expression. Mol Cell Biol. 2006;26:7901–7912. doi: 10.1128/MCB.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J Biol Chem. 1971;246:7131–7138. [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB. Origins and targets of translational control. In: Hershey JWB, Mathews MB, Sonenberg N, editor. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1996. pp. 1–29. [Google Scholar]