Abstract
Microtubule-stabilizing (MTS) agents, such as taxanes, are important chemotherapeutics with a poorly understood mechanism of action. We identified a set of genes repressed in multiple cell lines in response to MTS agents and observed that these genes are overexpressed in tumors exhibiting chromosomal instability (CIN). Silencing 22/50 of these genes, many of which are involved in DNA repair, caused cancer cell death, suggesting that these genes are involved in the survival of aneuploid cells. Overexpression of these “CIN-survival” genes is associated with poor outcome in estrogen receptor–positive breast cancer and occurs frequently in basal-like and Her2-positive cases. In diploid cells, but not in chromosomally unstable cells, paclitaxel causes repression of CIN-survival genes, followed by cell death. In the OV01 ovarian cancer clinical trial, a high level of CIN was associated with taxane resistance but carboplatin sensitivity, indicating that CIN may determine MTS response in vivo. Thus, pretherapeutic assessment of CIN may optimize treatment stratification and clinical trial design using these agents.
Keywords: chemotherapy, drug resistance
The clinical efficacy of microtubule-stabilizing (MTS) drugs is limited by intrinsic and acquired drug resistance. The mechanistic basis for taxane efficacy may be attributable to pathways downstream of a mitotic arrest that result in mitotic catastrophe, promoting cell death in metaphase or death preceded by multinucleation (1). A competent mitotic checkpoint and mitotic slippage plays a central role in cell death in response to an aberrant mitosis, kinesin spindle protein (KSP; kinesin-5 or Eg5) inhibition or paclitaxel exposure (2, 3). The molecular pathways resulting in taxane-induced cell death following mitotic checkpoint activation or death in response to an aberrant mitosis remain unclear, however. Sensitivity to microtubule-targeted drugs may depend on cellular pathways involved in maintaining chromosomal stability (4, 5). In this regard, the correlation of taxane resistance with increasing CIN in cancer cell lines may explain the in vivo taxane resistance of colorectal cancers, a disease with a high frequency of CIN (4, 6). CIN is associated with both poor prognosis in solid tumors and the rapid acquisition of multidrug resistance in cell culture models (7–9). CIN also is associated with altered cytotoxic response in vitro, providing a pharmacologically exploitable phenotype (10).
Microarray profiling of drug-induced gene expression changes has identified shared intracellular pathways through which disparate small molecules exert their cytotoxic activity (11–13). MTS drugs such as taxanes and epothilones, which compete for a similar microtubule-interacting region, initiate similar gene expression changes, suggesting that disruption of the microtubule network induces a characteristic gene expression response (14). Such gene expression changes may reflect the inhibition of transcription during mitosis, resulting in the degradation of short-lived mRNAs encoding antiapoptotic proteins, thereby modulating cell fate in response to microtubule stabilization (15).
Examining the regulation and function of transcripts that are consistently altered following MTS treatment may identify pathways leading to taxane-induced cell death and the survival of aneuploid cells. We reasoned that (i) MTS agents might trigger a cytotoxic gene expression program that prevents the survival of aneuploid progeny resulting from an aberrant mitosis; (ii) the initiation of this cytotoxic gene expression program in response to taxanes may be dysfunctional in CIN tumors, thus contributing to taxane resistance; and (iii) altered expression of genes implicated in the survival of CIN tumors may predict taxane sensitivity in vivo.
Results
Genes Repressed by MTS Are Overexpressed in CIN Tumors.
We derived a common MTS gene expression signature through a meta-analysis of published microarray data sets from 4 cancer cell lines and an ovarian cancer xenograft model treated with taxanes or epothilone [supporting information (SI) Fig. S1 and Table S1a] (14, 16, 17; GSE2182).
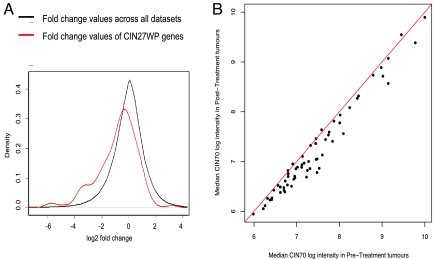
Using a list of published genes with elevated expression in high-CIN tumors (9), we tested whether genes repressed by MTS treatment tend to be overexpressed in tumors with high levels of CIN. To avoid nonspecific gene expression changes caused by the inhibition of cellular proliferation by MTS treatment, we excluded cell cycle–regulated genes from the CIN70 signature, leaving a signature of 27 genes (designated CIN27wp) (18). We compared the empirical frequency distribution of CIN27wp genes with the gene expression changes across all data sets following MTS exposure using a 1-sided bootstrap Kolmogorov-Smirnov test (Table S1b and Fig. 1A). We observed a significant left shift of the empirical frequency distribution of the CIN27wp genes (P = 3.3e-07), indicating that these genes are more likely to be repressed after MTS treatment. We also noted a significant enrichment of CIN genes in the MTS expression signature. This enrichment was not evident in the expression signature induced by a different cytotoxic agent, 5-FU, derived by similar methods (Fig. S2A and Table S1c), indicating that CIN gene repression is not a general response to cytotoxic agents.
Fig. 1.
Genes overexpressed in tumors with CIN are repressed by MTS treatment in vitro and in vivo. (A) The CIN27wp gene expression signature, which correlates with total functional aneuploidy in several cancer types (9), was analyzed in 5 gene expression data sets measuring response to MTS treatment. The distribution of changes induced by MTS was lower in the CIN27wp genes (red) compared with the set of all measured genes (black) (P = 3.3e-7). (B) As part of the OV01 clinical trial, expression profiling was performed on ovarian carcinomas before and after 3 cycles of paclitaxel (Px) treatment. For each gene in the CIN70 signature, the median expression in the pretreatment and posttreatment tumors was compared, with the red line indicating equivalence.
We also observed that genes overexpressed in CIN tumors are significantly repressed in taxane-sensitive ovarian cancer xenografts but less repressed in a paclitaxel-resistant xenograft model 24 h after treatment with 60 mg/kg of paclitaxel (Table S1d) (17). In addition, in the OV01 stage III-IV ovarian cancer clinical trial, most of the CIN70 and CIN27wp genes were repressed after 3 cycles of paclitaxel chemotherapy (Fig. 1B and data not shown). These results demonstrate that repression of genes associated with CIN occurs after taxane treatment in vivo. Consistent with the ovarian xenograft data, we observed significant repression of the median expression values of the CIN70 signature after paclitaxel therapy (Fig. S2 B–D), with a trend toward greater repression in the most paclitaxel-sensitive tumors (Fig. S2D). These data suggest that the repression of genes overexpressed in CIN tumors following taxane treatment may be functionally implicated in drug response.
Validation of MTS-Repressed Genes by Real-Time PCR.
We assessed whether the gene expression changes derived from our metasignature can be observed following MTS exposure of a cell line not used in the meta-analysis. We identified 50 MTS-repressed genes from our meta-analysis that also were overexpressed in CIN tumors (Fig. S3A) (9), and we randomly chose 36 of these genes for qPCR validation. We exposed the near-diploid HCT-116 colorectal cancer cell line to 50 nM paclitaxel and measured the response by qPCR. Expression of these 36 genes was consistently repressed following paclitaxel exposure, indicating that the meta-analysis methods reliably detected common MTS-induced gene expression changes independent of tumor type (Fig. S3B).
Identification of CIN-Survival Genes.
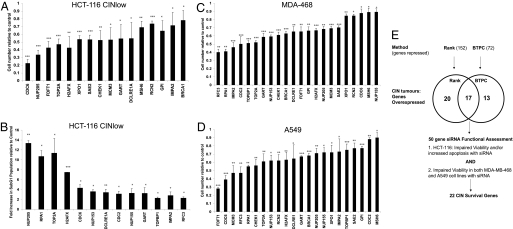
Because paclitaxel efficacy correlates with gene expression changes following treatment in xenografts (17), we predicted that the siRNA-induced silencing of MTS-repressed genes that are overexpressed in CIN tumors would lead to cytotoxicity in cancer cell lines. Indeed, silencing 22 of the 50 MTS-repressed genes significantly impaired cell viability and/or promoted a more than 2-fold increase in subG1 DNA content following gene silencing in the HCT-116 cell line in 3 independent experiments (Fig. 2 A and B). In addition, silencing all 22 of these genes significantly reduced cell viability in the A549 and MDA-MB-468 cancer cell lines, confirming a role for the expression of these genes in cancer cell survival in multiple tumor types (Fig. 2 C and D). Consistent with the contribution of aneuploidy induction after MTS treatment to cell death, 7/22 of these cell viability genes induced aneuploidy after RNAi-mediated silencing (19) (Fig. S4A). We validated the viability effects of 7 genes selected for follow-up by siRNA pool deconvolution (Fig. S4B) and confirmed gene silencing by qPCR (Fig. S4C), suggesting that these were not off-target effects.
Fig. 2.
Identification of 22 CIN-survival genes. (A) siRNA silencing of MTS-repressed genes impairs cell viability. Genes repressed within the 2 MTS expression signatures that are overexpressed in CIN tumors significantly altered HCT-116 cell viability when targeted by siRNA. An Acumen eX3 cytometer (Acumen TTP Labtech) was used to quantify viable cells 72 h after siRNA transfection. SDs are displayed for 3 independent experiments. P values are shown for Student 2-sided t-tests in all cases: *P < 0.05; **P < 0.005; ***P < 0.0005. (B) siRNA silencing of MTS-repressed genes promotes cell death. FACS analysis was used to quantify the mean subG1 fraction 72 h after transfection of siRNA. SDs and P values are displayed for 3 independent experiments. (C and D) Identification of 22 CIN-survival genes. Cells were transfected with siRNA targeting the 50 genes overexpressed in CIN tumors and repressed by MTS agents. Cell viability was quantified relative to scrambled control siRNA by a CellTiter-Blue assay 4 days after siRNA transfection in triplicate. Shown are the 22 genes that significantly impaired viability in the MDA-MB-468 and A549 cell lines. (E) Flowchart of analysis resulting in the derivation of 22 CIN-survival genes. The binomial test with probability corrections (BTPC) and rank methods were used to derive 2 MTS expression signatures. A total of 50 genes were repressed in the MTS signatures and overexpressed in CIN tumors. Of these 50 genes, 22 impaired cancer cell viability and/or induced apoptosis when silenced by RNA interference in 3 cancer cell lines: HCT-116 (colon), A549 (NSCLC), and MDA-MB-468 (breast).
In summary, we have functionally characterized the role of genes overexpressed in CIN tumors that are consistently repressed by MTS agents. We have identified 22 CIN-survival genes that are overexpressed in CIN tumors, repressed by MTS treatment, and impair cancer cell survival when depleted from 3 cancer cell lines of different tumor origin (Fig. 2E). Conceivably, repression of these genes may contribute to the cytotoxic response after MTS exposure.
Repression of CIN-Survival Genes in CINlow but Not CINhigh Cell Lines Correlates With Paclitaxel Cytotoxicity.
In this work, we defined CIN as a measure of cell-to-cell variability in chromosome number (20). We used published SKY data from NCI60 colorectal and breast cancer cell lines to define the fraction of normal chromosomes displaying numerical heterogeneity from cell to cell as a measure of CIN; we categorized cell lines as CINlow if <15% of chromosomes displayed numerical heterogeneity and as CINhigh if >40% of chromosomes exhibited numerical heterogeneity (21).
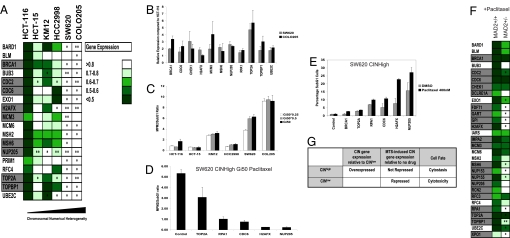
We reported previously that CIN correlates with resistance to paclitaxel in the NCI60 colon and breast cancer cell lines (6). To investigate whether the CIN status of a cell line correlates with the gene expression response following MTS treatment, we chose 6 NCI60 colon cancer cell lines with various degrees of CIN. We treated each cell line with a paclitaxel concentration equal to half of its Gi50 (drug concentration required for 50% growth inhibition; Table S2) for 24 h and measured gene expression by qPCR. Cell lines with the lowest frequency of CIN elicited repression of genes identified in the MTS signature, including several CIN-survival genes (Fig. 3A). In contrast, CINhigh cell lines (SW620 and COLO205) elicited significantly less repression of the CIN-survival genes compared with near-diploid CINlow HCT-116 cells. In agreement, greater repression of CIN-survival genes was observed in the CINlow MCF-7 breast cancer cell line compared with the CINhigh BT549 breast cancer cell line (Fig. S5A). Most of the CIN-survival genes exhibited increased basal expression in CINhigh cells compared with the CINlow HCT-116 cell line (Fig. 3B), in agreement with the published CIN signature (9).
Fig. 3.
Paclitaxel cytotoxicity is associated with CIN-survival gene repression. (A) Quantification of gene repression following paclitaxel treatment of cell lines with increasing chromosomal numerical heterogeneity. Fold change in gene expression post-paclitaxel treatment (24 h) relative to expression in vehicle control treated cells was determined by qPCR analysis (normalization to 18S and GAPDH) after 24 h of paclitaxel treatment (50% of the Gi50 concentration) in 3 biological replicate experiments in cell lines with increasing CIN. Color-coding represents the mean fold change in gene expression of 3 biological replicates relative to cells treated with vehicle alone + 1 SD. CIN-survival genes are highlighted in gray. Gene expression following paclitaxel treatment is compared with the HCT-116 cell line displaying the lowest CIN. The significance of the differences in gene expression were determined using the unpaired 2-sided Student t-test: *P <0.05; **P <0.005. (B) CIN-survival genes are relatively overexpressed in CINhigh COLO205 and SW620 cells compared with near-diploid CINlow HCT-116 cells. Shown is the relative quantification of gene expression normalized to 18S and GAPDH in COLO205 and SW620 cells compared with near-diploid HCT-116 cells. The graph indicates the mean fold change in gene expression compared with HCT-116 cells of 3 biological replicates (+ 1 SD). (C) Colorectal cancer cell lines with increasing chromosomal numerical heterogeneity uncouple mitotic arrest from cell death. Cell lines were treated with paclitaxel relative to the Gi50, Gi50/2, and Gi50/4 concentrations. Quantification of dying cells (subG1 fraction) and cells arrested in mitosis (MPM2-positive fraction) were assessed by FACS analysis. The MPM2:subG1 ratio was calculated by assessing the percentage of MPM2 positive cells relative to the percentage of subG1 cells following 24 h of paclitaxel treatment. The mean ratio of 3 independent experiments is presented (+1 SD). Cell lines are represented in ascending order of CIN status (21). (D and E) Silencing CIN-survival genes in the SW620 CINhigh cell line promotes cell death after 24 h of paclitaxel Gi50 treatment. At 48 h after transfection of CIN-survival siRNAs, SW620 cells were treated with paclitaxel for 24 h, and cells were prepared and analyzed as in C. In D, the mean MPM2:subG1 ratio of 2 independent experiments is presented (+ 1 SD). E shows that silencing of NUP205, H2AFX, CDC6, and RPA1 promoted a significant increase in subG1 cells after paclitaxel exposure compared with DMSO-treated cells (P <0.05; Student t-test). (F) Impaired repression of CIN-survival genes following paclitaxel treatment in an isogenic model of CIN. Quantification of gene repression by qPCR analysis of 21 CIN-survival genes following treatment of HCT-116 wild-type parental cells and isogenic Mad2+/- cells after 24 h of paclitaxel treatment (50 nM) normalized to 18s. Color-coding represents the mean fold repression + 1 SD from 3 biological replicates. P values indicate significantly greater gene repression in the HCT-116 parental cell line (Student 1-sided t-test). (G) CIN-survival gene repression correlates with cytotoxic response and stable tumor karyotype.
We suspected that the relative resistance of CINhigh cell lines to paclitaxel-mediated cell death may result from the failure of the drug to repress the CIN-survival genes. To investigate this, we tested whether the CIN status of the cell correlates with reduced cell death (based on the percentage of subG1 cells as determined by FACS) in the 6 colorectal cancer cell lines at different paclitaxel concentrations (Gi50/4, Gi50/2, and Gi50 concentrations for each cell line; Table S2). We observed a correlation between increasing CIN status of the 6 cell lines and an increased proportion of cells in mitosis relative to the percentage of dying cells (MPM2:subG1 ratio) after 24 h of paclitaxel treatment (at Gi50/4, Gi50/2, and Gi50 for each cell line; correlation coefficient, 0.80; 95% confidence interval, 0.77–0.86). The SW620 and COLO205 CINhigh cell lines that failed to efficiently repress the CIN-survival genes in Fig. 3A displayed the highest MPM2:subG1 ratios, consistent with a cytostatic rather than a cytotoxic response to drug exposure in CINhigh cells (Fig. 3C).
Next, we tested whether the silencing of CIN-survival genes increases paclitaxel-mediated cytotoxicity and lowers the MPM2:subG1 ratio in the CINhigh cell line SW620 (Fig. 3D). After silencing of NUP205, H2AFX, CDC6, RPA1, and TOP2A, we observed a significant decrease in the MPM2:subG1 ratio (with gene silencing confirmed by qPCR; data not shown). Silencing of NUP205, H2AFX, CDC6, and RPA1 promoted a significant increase in paclitaxel-induced cytotoxicity relative to vehicle-treated SW620 cells (Fig. 3E), supporting the model in which the repression of CIN-survival genes contributes to cytotoxicity following MTS exposure.
To substantiate the impaired repression of CIN-survival genes in CIN cells following taxane exposure, we investigated the repression of these genes in isogenic models of chromosomal instability. We noted significantly less repression of most of the CIN-survival genes tested in the HCT-116 Mad2+/− and early-passage HCT-116 hSecurin-/- after a 24-h exposure to 50 nM paclitaxel compared with parental control cells (Figs. 3F and S5B). These data support the impaired repression of CIN-survival genes in CIN tumor cells compared with the diploid isogenic pair after MTS exposure.
In summary, during taxane treatment, CINhigh cancer cell lines and isogenic CIN models did not repress CIN-survival genes as significantly as CINlow cell lines, and the CINhigh cell lines had greater basal expression of these genes. The cytotoxicity of paclitaxel is associated with robust repression of CIN-survival genes in CINlow cells (Fig. 3G). These data indicate that CIN may attenuate the cytotoxic response to MTS exposure through impaired CIN-survival gene repression. Consistent with this hypothesis, cytotoxicity in a CINhigh cell line can be partially restored by silencing CIN-survival genes.
CIN Ovarian Cancers Display Intrinsic Paclitaxel Resistance.
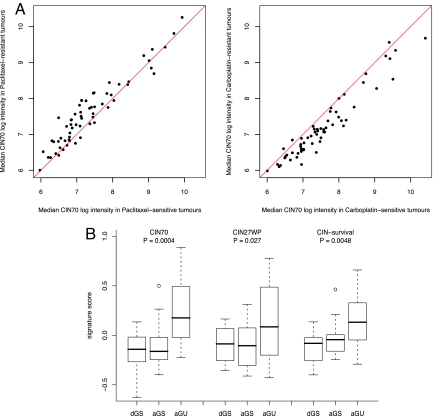
We used the OV01 clinical trial data set with objective response assessment to address whether CIN predicts sensitivity to paclitaxel in contrast to sensitivity to carboplatin in patients with ovarian cancer. We compared the median expression of each CIN70 gene in tumors before treatment with paclitaxel or carboplatin monotherapy and used established methods to define treatment response according to the fall in serum CA125 level (22). We found that most of the CIN70 genes were overexpressed in paclitaxel-resistant and carboplatin-sensitive tumors (Fig. 4A). High median CIN70 signature expression was associated with paclitaxel resistance, and low median CIN70 signature expression was associated with paclitaxel sensitivity (P = 0.043). CIN70 signature expression was significantly greater in tumors subsequently resistant to paclitaxel than in tumors resistant to carboplatin (P = 0.044), supporting the hypothesis that the efficacy of these 2 cytotoxic agents is differentially altered in CIN tumors. There was no significant relationship between Ki-67 or ABCB1, ABCB4, and ABCB11 expression and the response to paclitaxel, supporting the contribution of CIN rather than effects of tumor cell proliferation or drug efflux to paclitaxel resistance (data not shown). A strong correlation was seen between the median CIN-survival gene expression and median CIN70 expression in ovarian cancers derived from this cohort (R = 0.93; P < 0.0001).
Fig. 4.
CIN70 predicts paclitaxel sensitivity and is a surrogate for CIN in breast cancer. (A) Expression of CIN70 genes determines sensitivity to paclitaxel and carboplatin. The figure contrasts basal median gene expression for each CIN70 gene in tumors with differing responses to paclitaxel and carboplatin. Paclitaxel-resistant tumors exhibited a higher median log-intensity of the CIN70 signature compared with paclitaxel-sensitive tumors (P = 0.043). CIN70 gene expression differed significantly between tumors subsequently resistant to paclitaxel and tumors resistant to carboplatin (P = 0.044; Student 2-sided t-test). (B) Expression of CIN and CIN-survival genes was greater in the aGU breast cancers. DNA image cytometry was used to classify breast cancers as dGS, aGS, or aGU. Boxplots summarize the expression of the CIN70, CIN27wp, and CIN-survival signatures within each group. P values were calculated using the Student 2-tailed t-test.
Next, we examined whether expression of the CIN signature is increased in residual paclitaxel-resistant tumors compared with paclitaxel-sensitive tumors. We found significant overexpression of CIN70 genes in residual ovarian cancers classified as resistant based on Rustin's CA-125 criteria (defined as a CA-125 coefficient >-0.5) compared with sensitive tumors (P = 0.01) (Fig. S5C) that was of greater magnitude in taxane-resistant tumors compared with the tumors most sensitive to paclitaxel (defined as a CA-125 coefficient <-1; P = 8.30e-8) (Fig. S5C). These data suggest that there may be a selection pressure for sustaining CIN in paclitaxel-resistant tumors.
Our findings suggest that CIN, as defined by expression of the CIN signature, may predict subsequent resistance to paclitaxel and sensitivity to carboplatin in vivo. This provides clinical evidence implicating CIN in pretreatment tumors as a marker of taxane resistance in vivo and indicates that the efficacy of 2 common chemotherapy drugs may be differentially influenced by tumor karyotype.
Relationship of CIN-Survival Genes to Breast Cancer Molecular Subtype and Prognosis.
We addressed the relationship between clinical and pathological variables in breast cancer (for which taxane is a major component of treatment regimens), with expression of the CIN signature and CIN-survival genes in an effort to better identify patient cohorts that might selectively benefit from taxane therapy. We used DNA image cytometry methods to directly assess CIN in 44 primary breast cancers for which we had gene expression data available (GSE11901) (23). This technique is used to measure nuclear DNA content in clinical samples, including fine-needle aspirate cytology samples, and allows assessment of tumor ploidy status and tumor classification into genomically stable and unstable subtypes (24). The 44 breast cancers were classified into 3 groups: (i) aneuploid, genomically unstable breast cancers (aGU/CIN); (ii) aneuploid, genomically stable (aGS) tumors; and (iii) diploid, genomically stable (dGS) tumors. The aGU tumors have a wider distribution in image analysis-quantified DNA content relative to aGS or dGS tumors (Table S3). We assessed the expression levels of the CIN signature and CIN-survival genes in each of these tumors, classified for genomic instability status and clinicopathologic subtype by the median centroid method. The aGU tumors exhibited significantly greater expression of the CIN70 (P = 4e-04), CIN27wp (P = 0.027), and CIN-survival (P = 0.0024) gene signature set compared with the aGS and dGS tumors (Fig. 4B), confirming that expression of the CIN70 and CIN-survival signatures reflect CIN in vivo. The aGU tumors with the highest expression of the CIN and CIN-survival gene signatures represented 9 of 9 basal-like breast cancers and 4 of the 6 HER2-positive cancers in the 44-patient cohort.
We previously reported that the CIN70 expression signature has prognostic significance in breast cancer (9). Consistent with those earlier results, here the overexpression of CIN-survival genes correlated significantly with poorer disease-free and disease-specific survival (P < 0.05; log-rank test), with an increased hazard ratio for death or relapse seen in 3 of the 4 untreated estrogen receptor–positive breast cancer cohorts studied (Fig. S5D and Sweave analysis results).
In summary, our data indicate that CIN-survival gene expression is highly correlated both with the CIN signature in vivo and with CIN determined by DNA image cytometry. The CIN signature (9) and CIN-survival gene expression have prognostic power in breast cancer, and the CIN signature has predictive power and may serve as a surrogate marker for taxane resistance and carboplatin sensitivity in ovarian cancer.
Discussion
CIN-Survival Genes and Taxane Response.
Recent elegant work has classified the kinetics of cell death after MTS or K5I exposure in multiple cell lines, demonstrating the complexity and variation in this process within and between cell lines and challenging the prediction of tumor drug sensitivity in vivo (25, 26). In the present study, we explored how MTS-induced cytotoxicity might be orchestrated and how tumor response might be predicted in advance of treatment in vivo using cell population gene expression approaches.
Through this approach, we have identified a validated set of CIN-survival genes repressed by MTS agents that are significantly overexpressed in CIN tumors (9). Depletion of these genes appeared to induce cell death in both CINlow (HCT-116) and CINhigh (A549) cells (Fig. 2); however, CIN cell lines failed to repress CIN-survival genes after paclitaxel exposure as efficiently as CINlow cells, despite accumulating in mitosis in response to paclitaxel treatment, suggesting an uncoupling of mitotic arrest from cell death in CINhigh tumor cells associated with an attenuated cytotoxic gene expression program. These data support the robust mitotic arrest observed in chromosomally unstable cell lines after spindle disruption (27) and the lack of correlation between cell fate and the duration of mitotic arrest in time-lapse light microscopy studies (25, 26). Our findings also illustrate how MTS agents promote either a cytostatic or cytotoxic response that may correlate with the CIN status of the tumor (see the model shown in Fig. 3G). In agreement with this, CIN signature expression was correlated with resistance to paclitaxel and sensitivity to carboplatin in ovarian cancers, and the CIN signature was relatively overexpressed in residual, paclitaxel-resistant tumors. Thus, tumor karyotype may be an important determinant of cytotoxic sensitivity in human tumors.
Functional annotation of CIN-survival genes within the MTS expression signature may shed light on the mechanisms triggering cell death in response to MTS agents. Transcriptional inhibition during mitosis may alter the balance of short-lived mRNAs compared with long-lived mRNAs, initiating the degradation of mRNAs encoding proteins that promote cell survival (15). In agreement with this hypothesis, we found that several CIN-survival genes also were repressed following treatment with the transcription inhibitor flavopiridol (i.e., TOPBP1, CDC2, TOP2A, RFC3, and XPO1), indicating that cell death associated with MTS agents and transcriptional inhibitors may share similar mechanisms.
Recently it has been shown that mitotic arrest in response to paclitaxel triggers DNA damage (28), which may accumulate during an extended mitosis. Intriguingly, 9 of 22 genes in the CIN survival signature play a role in DNA repair. Repression of CIN-survival genes with roles in DNA repair after taxane exposure (e.g., BRCA1, CHEK1, DCLRE1A, H2AFX, MSH6, RPA1, RFC3, SAE2, and TOPBP1) suggests that failure to initiate efficient mismatch repair or homologous recombination may contribute to MTS cytotoxicity. It is noteworthy that these genes are overexpressed in CIN tumors and that mismatch repair deficiency is infrequent in CIN colorectal cancers, supporting the potential dependence of CIN on heightened DNA repair activity and a rational basis for the tumors' therapeutic resistance to MTS agents. Such a concept is supported by ploidy-specific lethal mutations in genes involved in homologous recombination in polyploid yeast models (29).
Clinical Trials Directed to Tumor Karyotype.
Using DNA image cytometry combined with gene expression analysis to classify cancers by genomic instability status, we have confirmed that the CIN signature is a surrogate marker of CIN in vivo and that CIN-survival genes are overexpressed in aneuploid, genomically unstable breast cancers. Furthermore, CIN signature expression appears to be greater in residual paclitaxel-resistant tumors than in sensitive tumors, suggesting that there may be a selection pressure for sustaining CIN in paclitaxel-resistant tumors.
Intriguingly, aGU tumors are almost exclusively basal-like, HER2-positive, luminal B breast cancers that, analogous to our clinical trial data in ovarian cancer, might be predicted to be relatively taxane-resistant but platinum-sensitive. HER2 expression confers resistance to taxanes that can be reversed by trastuzumab exposure (30), and in vitro studies of BRCA1-associated breast cancers (frequently associated with basal-like phenotype) have demonstrated sensitivity to platinum drugs and taxane resistance (31, 32). This has led to clinical trials assessing the efficacy of a taxane or a platinum agent in patients with HER2-negative, estrogen receptor–negative, and progesterone receptor–negative (so-called “triple-negative” breast cancers, which histologically often form basal-like tumors), or BRCA carrier metastatic breast cancers.
Unstable aneuploidy is associated with poor prognosis in cancer (9, 24) and may provide a catalyst to promote acquired drug resistance (7, 8). Here we have extended these previous observations, using a combination of functional genomic and microarray expression data sets, demonstrating that CIN is functionally associated with altered intrinsic tumor sensitivity to 2 distinct chemotherapy agents. These results suggest that the direct quantification of CIN and karyotypic state in tumors using FISH, DNA image cytometry, or FACS analysis should contribute to treatment stratification and to clinical trial design using these agents.
Materials and Methods
CTCR-OV01 Clinical Trial Analysis.
Ovarian carcinoma data were obtained from the CTCR-OV01 clinical trial, approved by the Cambridge Local Research Ethics Committee (33). Expression data were acquired on HGU133Av2 arrays (Affymetrix), reannotated using ENSEMBL (release 46)-derived custom CDF, and processed using Robust Multichip Average methodology without background correction.
DNA Image Cytometry.
A total of 44 surgically removed tumors (from the Cancer Center Karolinska Institutet, obtained in accordance with the guidelines of the local ethical review board) were used to create touch preparation slides for quantitative measurement of the nuclear DNA content. Tissues were snap-frozen until further processing with TRIzol reagent (Invitrogen) for DNA and RNA extraction. The staining procedure on Feulgen-stained touch preparation slides, internal standardization, tumor cell selection, and classification of genomic instability status were based on previously published methods (24).
Sweave File R Scripts.
Sweave files for the R language analyses of the MTS signature meta-analysis methods, the CTCR-OV01 analyses, and the distribution of the 3 gene expression signatures in the cohort of breast tumors classified by DNA image cytometry and cDNA expression analysis shown in Fig. 4B (GSE11901) are available online (available at http://www.dspace.cam.ac.uk/handle/1810/217842).
See SI Materials and Methods for more details on the methodology and for additional tables and figures.
Supplementary Material
Acknowledgments.
This work was funded by Cancer Research UK, the Medical Research Council, the National Institutes of Health (Grants NCI SPORE P50 CA 89393 and R21LM008823–01A1), and the Breast Cancer Research Foundation (Z.S.). C.S. is a Medical Research Council funded senior clinical research fellow. We thank Drs. Moreno, Bani, and Horwitz for microarray data sets; Professor Vogelstein and Drs. Jallepalli and Benezra for hSecurin-/- and Mad2+/− cells; and Professor Berres, Aengus Stewart, and Gavin Kelly for statistical support. We also thank the patients and the clinical staff at Addenbrookes and the Karolinska University Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811835106/DCSupplemental.
References
- 1.Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Tao W, et al. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer Cell. 2005;8:49–59. doi: 10.1016/j.ccr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 4.Swanton C, Tomlinson I, Downward J. Chromosomal instability, colorectal cancer and taxane resistance. Cell Cycle. 2006;5:818–823. doi: 10.4161/cc.5.8.2682. [DOI] [PubMed] [Google Scholar]
- 5.Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004;64:2502–2508. doi: 10.1158/0008-5472.can-03-2013. [DOI] [PubMed] [Google Scholar]
- 6.Swanton C, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Duesberg P, Stindl R, Hehlmann R. Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy. Proc Natl Acad Sci USA. 2000;97:14295–14300. doi: 10.1073/pnas.97.26.14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duesberg P, Stindl R, Hehlmann R. Origin of multidrug resistance in cells with and without multidrug resistance genes: Chromosome reassortments catalyzed by aneuploidy. Proc Natl Acad Sci USA. 2001;98:11283–11288. doi: 10.1073/pnas.201398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 10.Roschke AV, et al. Karyotypic “state” as a potential determinant for anticancer drug discovery. Proc Natl Acad Sci USA. 2005;102:2964–2969. doi: 10.1073/pnas.0405578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 12.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 13.Nunoda K, et al. Identification and functional signature of genes regulated by structurally different ABL kinase inhibitors. Oncogene. 2007;26:4179–4188. doi: 10.1038/sj.onc.1210179. [DOI] [PubMed] [Google Scholar]
- 14.Chen JG, Yang CP, Cammer M, Horwitz SB. Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res. 2003;63:7891–7899. [PubMed] [Google Scholar]
- 15.Blagosklonny MV. Mitotic arrest and cell fate: Why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6:70–74. doi: 10.4161/cc.6.1.3682. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Vargas H, Palacios J, Moreno-Bueno G. Molecular profiling of docetaxel cytotoxicity in breast cancer cells: Uncoupling of aberrant mitosis and apoptosis. Oncogene. 2007;26:2902–2913. doi: 10.1038/sj.onc.1210102. [DOI] [PubMed] [Google Scholar]
- 17.Bani MR, et al. Gene expression correlating with response to paclitaxel in ovarian carcinoma xenografts. Mol Cancer Ther. 2004;3:111–121. [PubMed] [Google Scholar]
- 18.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JG, Horwitz SB. Differential mitotic responses to microtubule-stabilizing and -destabilizing drugs. Cancer Res. 2002;62:1935–1938. [PubMed] [Google Scholar]
- 20.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining “chromosomal instability.”. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Roschke AV, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 22.Rustin GJ, et al. Re: New guidelines to evaluate the response to treatment in solid tumors (ovarian cancer) J Natl Cancer Inst. 2004;96:487–488. doi: 10.1093/jnci/djh081. [DOI] [PubMed] [Google Scholar]
- 23.Habermann JK, et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–1564. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronenwett U, et al. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–909. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- 25.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:1–12. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269–3276. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- 27.Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalton WB, Nandan MO, Moore RT, Yang VW. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67:11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storchova Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, et al. Overexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol Cell. 1998;2:581–591. doi: 10.1016/s1097-2765(00)80157-4. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 32.Quinn JE, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63:6221–6228. [PubMed] [Google Scholar]
- 33.Ahmed A, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitises ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.