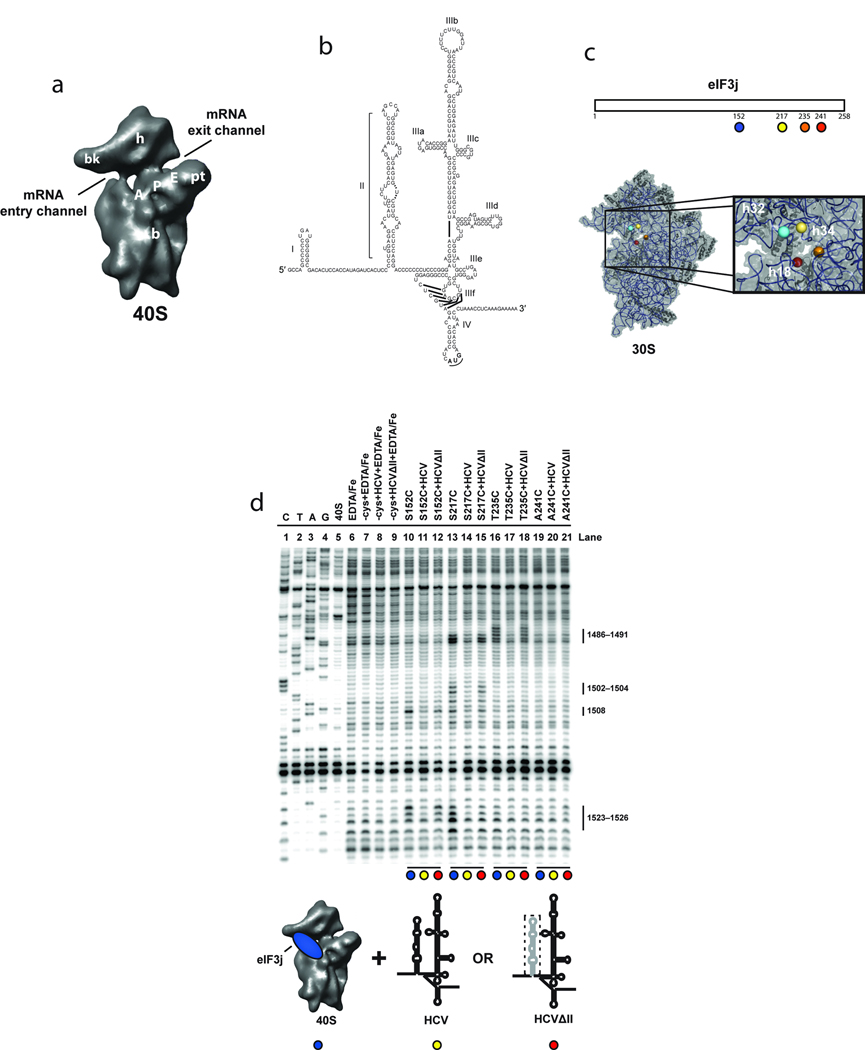
Figure 1.
Directed hydroxyl radical probing of 18S rRNA from BABE-Fe-eIF3j-40S-HCV complexes. (a) The 40S subunit structure based on a cryo-EM reconstruction13 viewed from the subunit interface with landmarks indicated: A, A-site; P, P-site; E, E-site; bk, beak; b, body; pt, platform; and h, head. (b) The 5´ UTR of the HCV mRNA consists of four domains (I–IV); the IRES domains (II–IV) with sub-domains (a–f) of domain III are indicated. (c) Representation of eIF3j indicating the positions of cysteine mutations used for BABE-Fe attachment (upper panel). Modeled positions of eIF3j amino acids in the T. thermophilus 30S crystal structure, adapted from a previous publication19 (lower panel). The boxed area provides a detailed view of the mRNA entry channel and A-site with helices 18, 32 and 34 indicated. Nucleotides cleaved in these helices for each experiment are shown in Supplementary Fig. 1a. (d) Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j. Sequencing lanes are indicated (C, T, A, and G). Other control lanes include 40S subunits in the absence or presence of EDTA/Fe, mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence (lane 7) or presence of wild type (HCV; lane 8), or domains III-IV (HCVΔII ; lane 9) of the HCV IRES RNA. Other lanes include eIF3j derivatized with BABE-Fe at the positions indicated either in the absence (lanes 10, 13, 16, and 19), or presence of HCV IRES (lanes 11, 14, 17, and 20), or HCVΔII IRES (lanes 12, 15, 18, and 21). 18S rRNA nucleotide positions of cleavage sites are indicated. Colored circles indicate components added in each reaction as depicted in the cartoon. The deletion of domain II (HCVΔII) is represented by a dotted line.