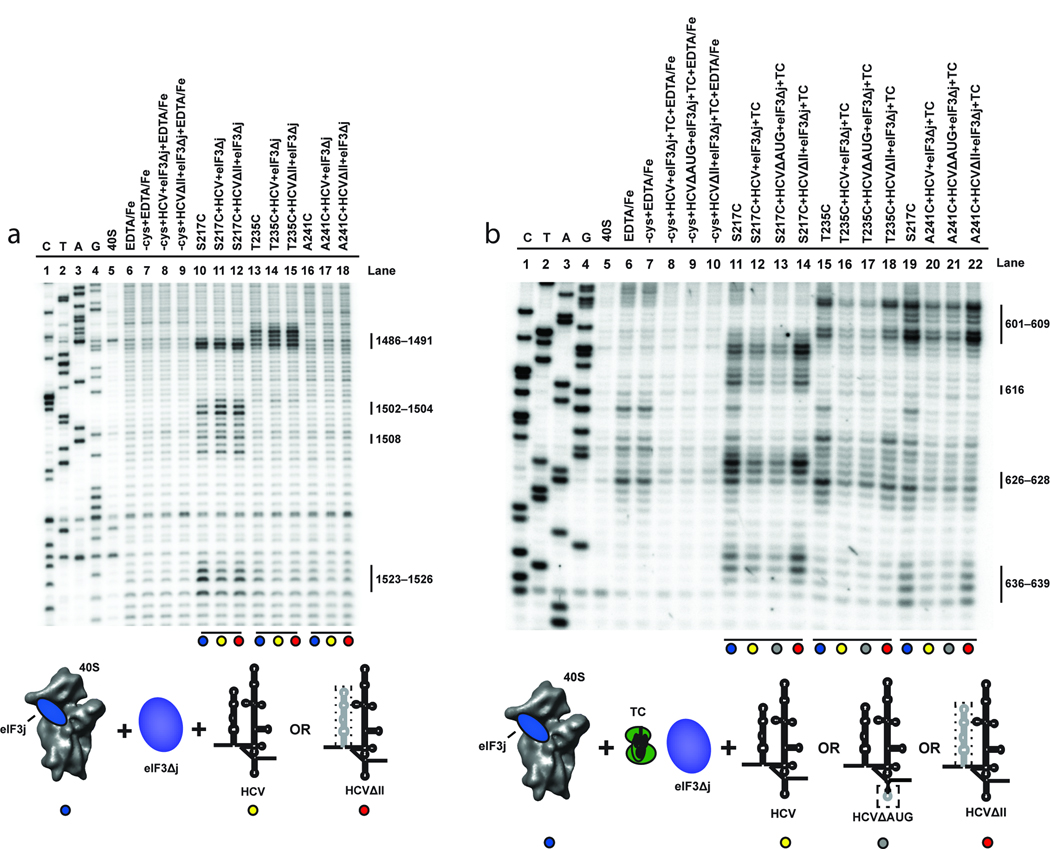
Figure 3.
Effects of eIF3 and eIF2-Met-tRNAi on directed hydroxyl radical probing of 18S rRNA with BABE-Fe-eIF3j. (a) Lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV constructs and eIF3 complex without endogenous eIF3j (eIF3Δj). Lanes corresponding to eIF3j derivatized with BABE-Fe at the positions indicated in the absence (lanes 10, 13, and 16), or presence of eIF3Δj and wild type HCV IRES (HCV; lanes 11, 14, and 17), or domain III of the HCV IRES (HCVΔII; lanes 12, 15, and 18) are indicated. (b) Lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV constructs and other initiation factors as indicated. Lanes corresponding to BABE-Fe-modified eIF3j at the positions indicated in the absence (lanes 11, 15, and 19) or presence of eIF3Δj, eIF2-Met-tRNAi (Ternary complex; TC) and HCV (lanes 12, 16, and 20), HCV\AUG (lanes 13, 17 and 21), or HCVΔII (lanes 14, 18, and 22) are indicated. For each gel, sequencing lanes (C, T, A, and G) and cleavage nucleotide positions in the 18S rRNA are indicated. Colored circles correspond to the components added, as depicted in the cartoons. Relevant mutations in each HCV IRES construct are represented by a dotted line.