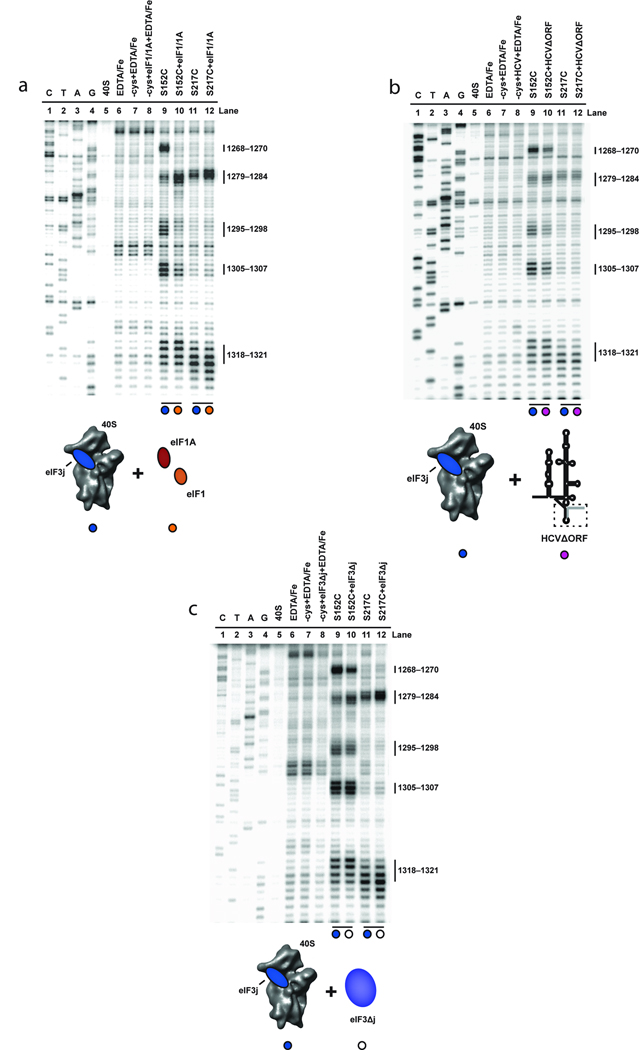
Figure 4.
Effects of eIF1, eIF1A, HCV and eIF3 and on directed hydroxyl radical probing of 18S rRNA from BABE-Fe-eIF3j. (a) Primer extension analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of eIF1 and eIF1A (lanes 10 and 12). (b) Analysis of 18S rRNA cleaved by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of HCVΔORF (lanes 10 and 12). (c) Analysis of 18S rRNA cleavage by BABE-Fe-modified eIF3j in the absence (lanes 9 and 11) or presence of eIF3Δj (lanes 10 and 12). In each gel the sequencing lanes are indicated (C, T, A, and G). Other lanes include 40S subunits in the absence or presence of EDTA/Fe and mock-derivatized eIF3j (−cys+EDTA-Fe) in the absence or presence of HCV and other initiation factors as indicated. Cleavage nucleotide positions in the 18S rRNA are indicated and colored circles correspond to the components added in each reaction, as depicted in the cartoons.