Abstract
The changes in cancer cell surface molecules and intracellular signaling pathways during tumorigenesis make delivery of adenovirus-based cancer therapies inefficient. Here we have identified carcinoembryonic antigen–related cell adhesion molecule 6 (CEACAM6) as a cellular protein that restricts the ability of adenoviral vectors to infect cancer cells. We have demonstrated that CEACAM6 can antagonize the Src signaling pathway, downregulate cancer cell cytoskeleton proteins, and block adenovirus trafficking to the nucleus of human pancreatic cancer cells. Similar to CEACAM6 overexpression, treatment with a Src-selective inhibitor significantly reduced adenovirus replication in these cancer cells and normal human epithelial cells. In a mouse xenograft tumor model, siRNA-mediated knockdown of CEACAM6 also significantly enhanced the antitumor effect of an oncolytic adenovirus. We propose that CEACAM6-associated signaling pathways could be potential targets for the development of biomarkers to predict the response of patients to adenovirus-based therapies, as well as for the development of more potent adenovirus-based therapeutics.
Introduction
Adenovirus-based cancer therapies hold promise for treatment of malignancies resistant to conventional therapy. The nonreplicating Ad-p53 and the replication-selective oncolytic adenovirus (RSOA) H101 have both been approved for head and neck cancer therapy by the Chinese State Food and Drug Administration (1), and both therapeutics have shown encouraging results when combined with chemotherapy (2, 3). Unfortunately, adenoviral monotherapy has demonstrated only limited efficacy (4–6). In addition, several tumor types, including pancreatic cancer, are poorly responsive to RSOA (7–9), suggesting that the genetic variability between tumors may play a key role in determining the infectivity of adenovirus vector. Genetic alterations underlying tumorigenesis can lead to changes in cell-surface molecules and intracellular signaling pathways that might affect the ability of adenovirus to infect and replicate efficiently in cancer cells.
There is increasing evidence that cellular signal transduction pathways affect adenovirus infection. Adenovirus endocytosis via αv integrins requires phosphoinositide-3-OH kinase and actin cytoskeleton reorganization mediated by Rho family GTPases (10, 11); cell signaling pathways are required not only for adenovirus vector cell entry, but also for subsequent intracellular trafficking and viral replication; for example, PKA is required for effective nuclear targeting of adenovirus, and p38-enhanced nuclear targeting of adenovirus is dependent on the downstream MAPK-activated protein kinase 2 (MK2) (12). The adenovirus-induced oncogenic Raf/MEK/ERK signaling pathway enhances viral progeny by sustaining the levels of viral proteins (13).
A well-known factor limiting the efficacy of adenovirus-based therapy has been shown to be paucity of expression of the Coxsackie and adenovirus receptor (CAR) on some tumors (4, 5, 14). While binding and endocytosis of adenoviral vectors into target cells are both necessary, they are not sufficient for successful gene delivery (15). Successful adenoviral infection also requires the ability to elicit membrane ruffling, macropinosome formation, effective trafficking to the nucleus, replication, lysis, and the induction of a migratory phenotype (16). These depend on the interaction of the virus and host cells. It has become evident that intracellular genetic alterations of tumor cells play an important role in the viral infection. The identification of cellular components affecting the life cycle of adenovirus will open up a new avenue to enhance the efficacy of adenovirus-based therapeutics. To this end, we screened a panel of pancreatic cancer cell lines for sensitivity to adenovirus and then examined differential gene expression between adenovirus-sensitive and -insensitive cells by Affymetrix array analysis. Interestingly, we found that expression of carcinoembryonic antigen–related cell adhesion molecule 6 (CEACAM6), a gene with a well-documented role in tumor invasion, metastasis, and drug resistance (17–20), correlates with the sensitivity of tumor cells to adenovirus. Further stepwise investigations demonstrated that CEACAM6 blocked adenovirus trafficking to the nucleus through the Src pathway, which interferes with the cytoskeleton of cancer cells, resulting in attenuated infectability by adenovirus. siRNA-mediated knockdown of CEACAM6 significantly increased adenovirus replication and antitumor efficacy of oncolytic adenovirus in vivo.
Results
A suitable cell model for screening tumor-associated genes affecting the potency of oncolytic adenovirus.
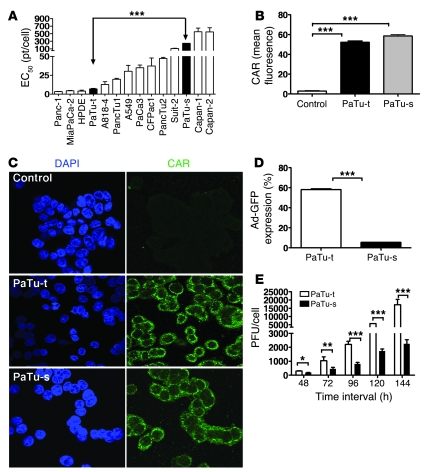
Given the poor response of human pancreatic cancer patients to oncolytic adenovirus, we first screened the sensitivity of 13 human pancreatic cell lines to replicating adenovirus. Different tumor cell lines showed variable responses to oncolytic adenovirus (Figure 1A). PaTu8988t and PaTu8988s cells derived from the same patient showed significantly different responses to adenovirus. In order to investigate the underlying mechanism, we analyzed CAR expression and adenovirus infectivity using an anti-CAR antibody (Figure 1, B and C) and nonreplicating Ad-CMV-GFP virus (Figure 1D) by FACS and immunocytochemistry. While higher GFP expression in PaTu8988t than in PaTu8988s cells was consistent with its greater sensitivity to the downstream oncolytic effects of this virus, CAR expression was actually lower in adenovirus-sensitive PaTu8988t cells compared with adenovirus-insensitive PaTu8988s cells. Given that replication of adenovirus in PaTu8988t was greater than in PaTu8988s cells (Figure 1E), it is likely that other genetic changes beyond low CAR expression are responsible for the different outcomes of adenovirus infection observed between these 2 cell lines, which represent a suitable model to discover such genes.
Figure 1. Sensitivity of human cancer cell lines to adenovirus; and CAR expression in cancer cells.
(A) Sensitivity of human pancreatic cell lines and the A549 lung cancer cell line to oncolytic adenovirus Ad5. EC50 (virus dose required to kill 50% of cells) values were derived by MTS assay. PaTu-t, PaTu8988t; PaTu-s, PaTu8988s. (B) FACS analysis showing the expression of adenovirus receptor CAR in PaTu8988t and PaTu8988s human pancreatic cancer cell lines. (C) The expression of adenovirus receptor CAR in PaTu8988t and PaTu8988s human pancreatic cancer cell lines by immunocytochemistry. Original magnification, ×600. (D) FACS analysis showing the infectivity of adenovirus Ad-GFP in human pancreatic cancer cell lines PaTu8988t and PaTu8988s infected with Ad-CMV-GFP adenovirus at an MOI of 100 pt/cell. (E) Adenovirus replication in human pancreatic cancer cell lines PaTu8988t and PaTu8988s (infected at an MOI of 100 pt/cell) assayed by the limiting dilution assay as previously described (41). All experiments were repeated at least 3 times. *P < 0.05, **P < 0.01, ***P < 0.001.
CEACAM6 is a tumor-associated gene that correlates with the sensitivity of pancreatic cancer cells and colorectal cancer cells to adenovirus.
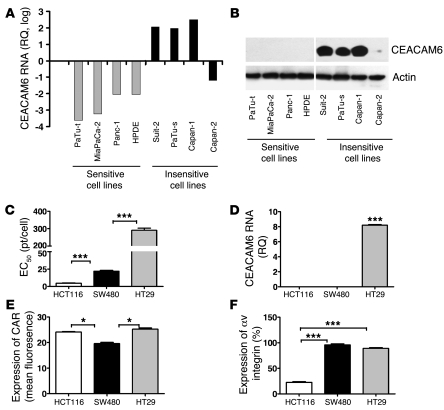
To identify those intrinsic tumor genes that potentially affect the potency of oncolytic adenovirus, the Affymetrix GeneChip (U133 Plus 2.0) was used to compare the gene expression profile of PaTu8988t and PaTu8988s cells. The gene expression profile of PaTu8988s and PaTu8988t is shown in Supplemental Table 1 (supplemental material available online with this article; doi: 10.1172/JCI37905DS1). CEACAM6 is one of the top 11 genes differentially expressed between the 2 cell lines, showing 89-fold higher expression in the PaTu8988s cell line (adenovirus-insensitive) compared with the PaTu8988t cell line (adenovirus-sensitive) (P < 0.001). Given the well-documented functions of CEACAM6 in cancer, we chose this gene as the first target to investigate further. In order to validate the correlation of CEACAM6 expression with virus sensitivity, the 4 most adenovirus-sensitive cell lines (Panc-1, MiaPaCa-2, HPDE, and PaTu8988t) and the 4 most insensitive cell lines (EC50 >100 particles/cell [pt/cell]; Suit-2, PaTu8988s, Capan-1, and Capan-2) in the panel were investigated for CEACAM6 expression by quantitative real-time PCR (qPCR). As shown in Figure 2A, in 7 of 8 human pancreatic cell lines examined, CEACAM6 expression correlates inversely with the sensitivity to adenovirus (r2 = 0.9112, P = 0.0004). CEACAM6 protein was undetectable in all sensitive cell lines but expressed at high levels in 3 of 4 adenovirus-insensitive cells (Figure 2B), consistent with mRNA expression.
Figure 2. CEACAM6 expression in cancer cells and adenovirus infectivity.
(A) Expression of CEACAM6 RNA in adenovirus-sensitive and -insensitive pancreatic cell lines as assayed by qPCR. The A549 cell line was used as a calibrator to calculate the relative quantity (RQ) values. (B) Expression of CEACAM6 protein in adenovirus-sensitive and -insensitive pancreatic cell lines as detected by Western blot analysis. (C) Sensitivity of colon cancer cell lines to adenovirus. EC50 values were derived by MTS assay as described above. (D) qPCR analysis showing the expression of CEACAM6 RNA in colorectal cancer cell lines. The A549 cell line was used as a calibrator to calculate the relative quantity values. (E) Expression of adenovirus receptor CAR in human colorectal cancer cell lines as analyzed by FACS. (F) Expression of integrin αv in human colorectal cancer cell lines as analyzed by FACS. All experiments were repeated at least 3 times. *P < 0.05, ***P < 0.001.
To determine whether the relationship between CEACAM6 expression and sensitivity to adenovirus existed in other tumor types, 3 colorectal cancer cell lines were investigated in a proof-of-principle experiment. The sensitivity of these 3 cell lines to adenovirus (Figure 2C) also inversely correlated with the level of CEACAM6 expression (Figure 2D). Only HT29, the cell line least sensitive to adenovirus, expressed a high level of CEACAM6. The sensitivity of the 3 cell lines to adenovirus was not related to the expression of CAR and integrin (Figure 2, E and F). These results suggest that CEACAM6 expression is inversely correlated with the sensitivity of cancer cells to adenovirus. CEACAM6 may be a useful biomarker for predicting the response of tumor cells to adenovirus-based gene therapy.
Overexpression of CEACAM6 in adenovirus-sensitive cancer cells reduces their infectability by adenovirus.
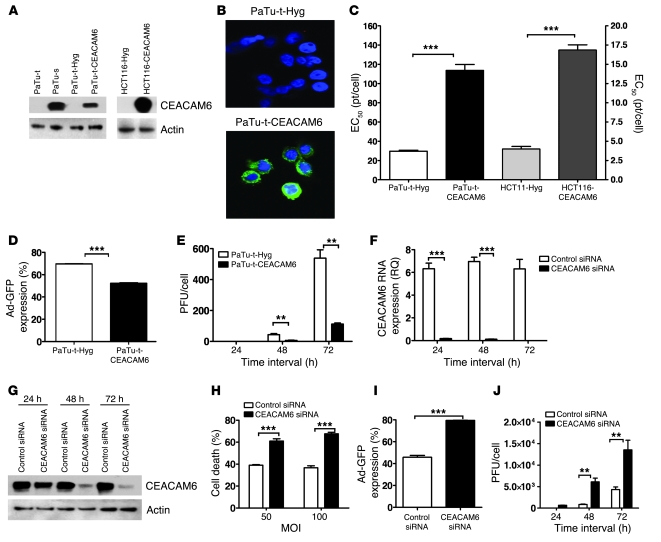
To assess the functional significance of elevated CEACAM6 expression for adenovirus infection, we investigated whether stable overexpression of CEACAM6 in cancer cell lines could alter their infectability by adenovirus. For this purpose, stable CEACAM6-expressing cell lines PaTu8988t-CEACAM6 and HCT116-CEACAM6 (neither of the parental lines of these clones expresses CEACAM6) and empty vector–transfected control cells were established by transfection with pML1-CEACAM6 and pML1-Hyg vectors, respectively, in which CEACAM6 expression was induced by zinc sulfate (17). CEACAM6 expression in the stable cell lines was verified by Western blot analysis and immunofluorescence staining (Figure 3, A and B). Intriguingly, overexpression of CEACAM6 led to more than 4-fold reduction in sensitivity to adenovirus in PaTu8988t and HCT116 cells compared with the vector-transfected cell lines (Figure 3C). Consistent with this, adenoviral infectivity and replication in CEACAM6-overexpressing PaTu8988t cells was also significantly reduced (Figure 3, D and E). These results demonstrate that CEACAM6 overexpression reduces the infectability of tumor cells by adenovirus. In order to further validate the effect of CEACAM6 on adenovirus infection, rescue experiments were performed to confirm that knocking down CEACAM6 expression in the cells that are transfected to overexpress CEACAM6 can restore the phenotype of the parental line. As shown in Supplemental Figure 1A, CEACAM6 expression in the PaTu8988t-CEACAM6 stable cell line was downregulated either by CEACAM6-specific siRNA or by withdrawal of zinc sulfate. The adenovirus infectivity (Supplemental Figure 1B) and cytotoxicity (Supplemental Figure 1C) in these 2 conditions were significantly rescued.
Figure 3. CEACAM6 functionally affects adenovirus infection.
(A) Expression of CEACAM6 protein in pancreatic cancer cell lines and engineered subclone cell lines as analyzed by Western blotting. (B) CEACAM6 expression in vector-transfected PaTu8988t (top) and CEACAM6-transfected PaTu8988t cell lines (bottom) by confocal microscopy, showing CEACAM6 expression on membrane and in cytoplasm. Blue (DAPI staining) indicates nuclei; green, CEACAM6; original magnification, ×600. (C) Sensitivity of CEACAM6-overexpressing cell lines and their counterparts to adenovirus as assayed by MTS. The EC50 values were increased 4-fold in CEACAM6-transfected PaTu8988t and HCT116 cells compared with their counterparts. (D) The infectivity of adenovirus Ad-GFP in stable clones of PaTu8988t-Hyg and PaTu8988t-CEACAM6 by FACS analysis at 48 hours after infection with Ad-CMV-GFP adenovirus. (E) Adenovirus replication in stable clones of PaTu8988t-Hyg and PaTu8988t-CEACAM6 (infected at an MOI of 100 pt/cell). (F) The expression of CEACAM6 as analyzed by qPCR in Suit-2 cell line after treatment with control siRNA and the CEACAM6-specific SMARTpool at various time points. (G) The expression of CEACAM6 protein by Western blotting after treatment with control siRNA, and the CEACAM6-specific SMARTpool at various time points. (H) Cell death of control siRNA– and CEACAM6-specific SMARTpool siRNA–pretreated Suit-2 cells after adenovirus infection at MOI of 50 and 100 pt/cell. (I) The infectivity of adenovirus Ad-GFP in control and CEACAM6-specific SMARTpool siRNA–pretreated Suit-2 cells by FACS at 48 hours after infection with Ad-CMV-GFP adenovirus. (J) Adenovirus replication in control and CEACAM6-specific SMARTpool siRNA–pretreated Suit-2 cells (infected at an MOI of 100 pt/cell) by 50% tissue culture infective dose (TCID50) assay. All experiments were repeated at least 3 times. **P < 0.01, ***P < 0.001.
Downregulation of CEACAM6 by siRNA enhances the infectability of cancer cells by adenovirus.
In order to further validate the effect of CEACAM6 on adenovirus infection and explore an approach to enhancing the sensitivity of tumor cells to adenovirus by targeting CEACAM6, we investigated whether downregulation of CEACAM6 by siRNA could enhance the infectability of tumor cells by adenovirus. Given that Suit-2 cells inherently express high levels of CEACAM6 and have a higher transfectability for delivery of siRNAs compared with PaTu8988s cells, this cell line was chosen to evaluate the effects of CEACAM6-specific siRNA on adenovirus infection. After optimization, we confirmed that CEACAM6 SMARTpool siRNAs almost completely suppressed CEACAM6 expression in Suit-2 cells at 72 hours after transfection (Figure 3, F and G). The control siRNA did not affect cell viability and apoptosis induction at the concentration used in the present study (data not shown). We then tested whether downregulation of CEACAM6 by siRNA could enhance the cytotoxicity of adenovirus in tumor cells endogenously expressing CEACAM6. Suit-2 cells were transfected with CEACAM6-specific siRNA or control siRNA for 72 hours prior to infection with wild-type adenovirus for a further 72 hours. The amount of cell death of Suit-2 cells after adenovirus infection at MOI of 50 and 100 increased at least 2-fold following pretreatment with CEACAM6-specific siRNA when compared with control siRNA (Figure 3H), consistent with the higher adenovirus infectivity (Figure 3I) and viral replication (Figure 3J). These results demonstrate that downregulation of CEACAM6 by siRNA can enhance the infectivity of adenovirus vector and the potency of oncolytic adenovirus. We therefore propose that CEACAM6-specific siRNA is a potential therapeutic to enhance adenovirus-based therapies. To validate this, we investigated whether restoring CEACAM6 expression after it has been knocked down by siRNA could restore the phenotype of cancer cells to adenovirus. As shown in Supplemental Figure 2, transfection of a CEACAM6-expressing plasmid into siRNA-knockdown Suit-2 cells elicited rescue of CEACAM6 expression (even if not to 100%) and indeed rescued the phenotype of adenovirus infectivity (Supplemental Figure 2B). In a time-course experiment (from day 3 to day 10 after transfection of CEACAM6 siRNA into Suit-2 cells), we found that the adenovirus infectivity (measured by GFP expression) and cytotoxicity in Suit-2 cells were rescued with the recovery of CEACAM6 expression over the time course after siRNA transfection (Supplemental Figure 2, C–E).
CEACAM6 inhibits the adenovirus life cycle by blocking adenoviral cytoplasmic trafficking.
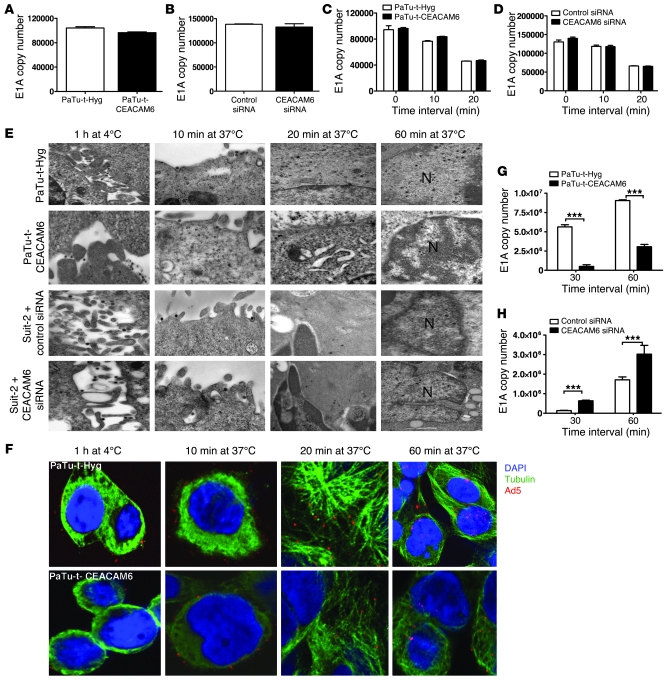
Since CEACAM6 expression affects the function of the nonreplicating Ad-GFP virus (Figure 1D and Figure 3, D and I), we hypothesized that CEACAM6 might affect phases of adenovirus infection between attachment and early gene expression. We therefore assessed the effect of CEACAM6 on adenovirus attachment, internalization, and movement of viral DNA to the nucleus in tumor cells. As demonstrated in Figure 1, B and C, CEACAM6 seems not to have affected expression of the adenovirus receptor CAR, an observation further confirmed by the fact that no difference in CAR expression was found between cells engineered to express CEACAM6 and those in which downregulation was achieved by CEACAM6-siRNA, when compared with their respective controls (Supplemental Figure 3). qPCR with E1A region primers, as previously described (21), was used to determine whether CEACAM6 affects the attachment of adenovirus (Figure 4, A and B). No difference in adenovirus attachment was seen between CEACAM6-negative cells (PaTu8988t-Hyg) and CEACAM6-expressing cells (PaTu8988t-CEACAM6) or between cells in which CEACAM6 was downregulated by siRNA and their counterparts. Next, we investigated whether CEACAM6 affects adenovirus internalization, since it has been reported that CEACAM6 can regulate integrin expression (22), which in turn may interfere with adenovirus internalization. To this end, CEACAM6-overexpressing or -downregulated cell lines and their controls were incubated with adenovirus for 60 minutes at 4°C. Unbound viruses were removed by washing with ice-cold PBS and cells warmed at 37°C for 10 and 20 minutes prior to the removal of uninternalized adenovirus by subtisilin. The absolute DNA copy number of adenovirus released by subtisilin from cell membrane detected by qPCR (Figure 4, C and D) was similar in CEACAM6 expression–modulated cells and control cell lines. This suggests that CEACAM6 does not affect the internalization of adenovirus, even though CEACAM6 affected the expression of integrin αvβ3 (Supplemental Figure 4). Since CEACAM6 expression affected the adenovirus infectivity as assayed by nonreplicating GFP virus (Figure 3, D and I), we hypothesized that CEACAM6 might affect adenovirus trafficking after internalization. To examine this, we used transmission electron microscopy (TEM) and confocal microscopy to monitor virus trafficking in the 4 CEACAM6 expression–modulated cells. CEACAM6 overexpression blocked adenovirus trafficking after internalization (Figure 4, E and F), resulting in decreased viral entry to the nucleus. In order to confirm this conclusively and quantitatively, after Ad5 was allowed to attach during incubation at 4°C for 1 hour and then allowed to traffic intracellularly by incubation at 37°C for 30 and 60 minutes, we determined the absolute DNA copy number of Ad5 E1A gene in the nucleus in each of these 4 cell lines. As shown in Figure 4, G and H, overexpression of CEACAM6 significantly reduced adenoviral entry to nuclei (Figure 4G), while downregulation increased it (Figure 4H).
Figure 4. Effect of CEACAM6 on the life cycle of adenovirus.
(A) Quantitation of E1A copy number after Ad5 binding in PaTu8988t-Hyg and PaTu8988t-CEACAM6 cells at 4°C for 1 hour (P = 0.5679) by qPCR. (B) qPCR quantitation of E1A copy number after Ad5 binding in control and CEACAM6-specific SMARTpool siRNA–pretreated Suit-2 cells at 4°C for 1 hour (P = 0.3619). (C) qPCR quantitation of E1A copy number of uninternalized adenovirus in PaTu8988t-Hyg and PaTu8988t-CEACAM6 cells (P > 0.05) at different time points. (D) qPCR quantitation of E1A copy number of uninternalized adenovirus in control and CEACAM6-specific SMARTpool siRNA–treated Suit-2 cells (P > 0.05) at different time points. (E) Adenovirus attachment and trafficking observed by TEM in CEACAM6-modulated cells and their counterparts after Ad5 was allowed to bind at 4°C for 1 hour and allowed to internalize and traffic to the nuclei at 37°C for various time points. N, nucleus. Original magnification, ×60,000. (F) Confocal images of stable cells with labeled Ad5 particle (red) and α-tubulin (green) after Ad5 was bound at 4°C for 1 hour and allowed to internalize and traffic to nuclei at 37°C for various times; original magnification, ×600. (G) Quantitation of E1A copy number in the nucleus after Ad5 was bound at 4°C for 1 hour and allowed to traffic at 37°C for 30 and 60 minutes in PaTu8988t-Hyg and PaTu8988t-CEACAM6 cells by qPCR. (H) qPCR quantitation of E1A copy number in the nuclei after Ad5 was allowed to bind at 4°C for 1 hour and allowed to traffic at 37°C for 30 and 60 minutes in control and CEACAM6-specific siRNA–treated Suit-2 cells by qPCR. ***P < 0.001.
CEACAM6 interferes with the Src pathway to disturb the cytoskeleton of cancer cells.
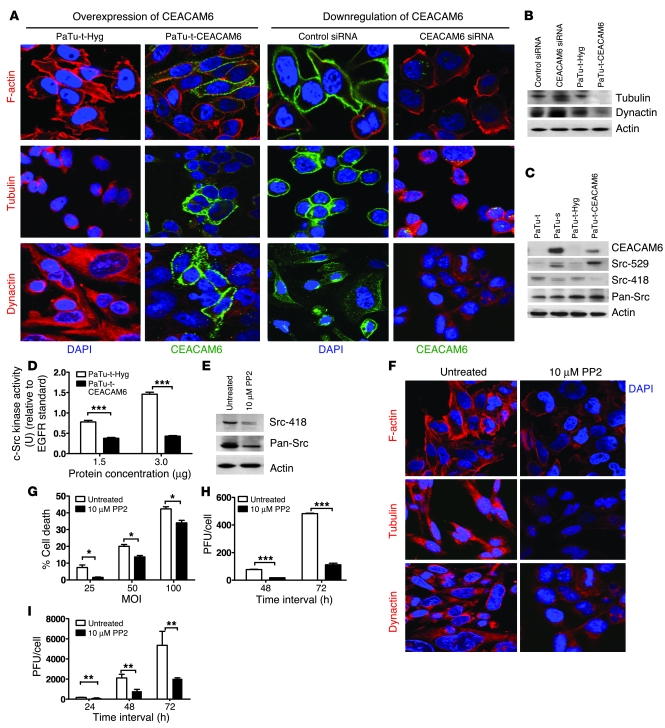
Cytoskeletal network proteins such as actin and tubulin act as “highways” to facilitate the trafficking of viral particles to specific destinations (23). Microtubules play an important role in adenovirus trafficking, and intact microtubules are necessary for successful adenoviral infection (24). Given that CEACAM6 affects adenovirus trafficking, we investigated whether CEACAM6 affected viral trafficking by interfering with the cytoskeleton. The expression of CEACAM6, F-actin, tubulin, and dynactin was detected by immunofluorescence in the CEACAM6 expression–modulated cells (Figure 5A). Overexpression of CEACAM6 altered the organization of cytoskeleton proteins and reduced tubulin and dynactin expression, as confirmed by immunoblotting (Figure 5B). In contrast, downregulation of CEACAM6 expression in Suit-2 cells by siRNA increased assembly of cytoskeleton proteins and increased expression of tubulin and dynactin (Figure 5, A and B).
Figure 5. CEACAM6 interferes with adenovirus infection by altering cytoskeleton of cancer cells via the Src pathway.
(A) Confocal analysis showing the expression of F-actin, tubulin, and dynactin in PaTu8988t-Hyg, PaTu8988t-CEACAM6, and control siRNA–treated Suit-2 cells and in CEACAM6-specific SMARTpool siRNA–treated Suit-2 cells; original magnification, ×600. DAPI (blue) indicates nuclei; green indicates CEACAM6; and red indicates F-actin, tubulin, or dynactin. (B) Western blot analysis showing the expression of tubulin and dynactin. (C) Western blot analysis showing the expression of CEACAM6, Src-529, Src-418, and pan-Src. (D) c-Src kinase activity in stable clones PaTu8988t-Hyg and PaTu8988t-CEACAM6. (E) Western blot analysis showing the expression of Src-418 and pan-Src in untreated and PP2-treated PaTu8988t cells. (F) Confocal analysis showing the expression of F-actin, tubulin, and dynactin in untreated and PP2-treated PaTu8988t cells; original magnification, ×600. (G) Cell killing assays with different MOI of adenovirus in untreated and PP2-pretreated PaTu8988t cells. (H) Adenovirus replication in untreated and PP2-pretreated PaTu8988t cells. (I) Adenovirus replication in untreated and PP2-pretreated normal human bronchial epithelial (NHBE) cells. All experiments were repeated at least 3 times. *P < 0.05, **P < 0.01, ***P < 0.001.
To explore the molecular mechanism by which CEACAM6 interferes with the cytoskeleton, we examined the effect of CEACAM6 on the Src pathway, since it is a major pathway involved in cytoskeleton regulation also reported to be affected by CEACAM6 expression (22). Total Src, phosphorylated Src, and Src activity were compared in CEACAM6-overexpressing cells and their counterparts. Surprisingly, we found that CEACAM6 overexpression in adenovirus-sensitive PaTu8988t cancer cells led to a reduction in phosphorylation of Src on T418 (active site) and an increase in phosphorylation of Src on T529 (Figure 5C), with accompanying decreased activity of Src (Figure 5D). In contrast, downregulation of CEACAM6 in adenovirus-insensitive Suit-2 cells by siRNA led to an increase in Src activity (data not shown). To confirm further that the CEACAM6-induced decrease in Src activity reduced adenovirus infection by disturbing the cytoskeleton, we used the Src-selective inhibitor PP2 to block Src function and then examined the effect of PP2 on cytoskeleton. PP2 treatment resulted in decreased Src activity by reducing the phosphorylation of Src at T418 (Figure 5E) and reduced assembly of cytoskeletal proteins in PaTu8988t cells (Figure 5F). Pretreatment of PaTu8988t cells with 10 μM PP2 also resulted in reduced sensitivity to and replication of adenovirus (Figure 5, G and H). This suggests that the Src pathway affects adenovirus infection through its action on the cell cytoskeleton. In addition, Src-selective inhibitors may have the potential to employed in antiviral therapy. Given that bronchial epithelium is known to be permissive to adenovirus infection, normal human bronchial epithelial (NHBE) cells were pretreated with PP2 and then infected with wild-type Ad5. PP2 dramatically inhibited adenovirus replication in NHBE cells (Figure 5I).
Systemic delivery of CEACAM6-specific siRNA enhances the antitumor potency of oncolytic adenovirus in vivo.
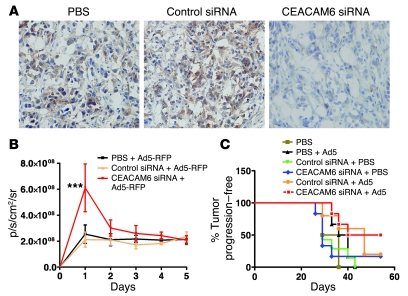
First, we determined whether the systemic delivery of siRNA by i.p. injection could reduce CEACAM6 expression and enhance viral replication in a subcutaneous Suit-2 xenograft tumor model (25). Delivery of CEACAM6-specific siRNA by i.p. injection daily for 3 days suppressed CEACAM6 expression in Suit-2 tumors in nude mice (Figure 6A). Treatment with control siRNA relative to PBS had no effect on CEACAM6 expression. To assess the effect of CEACAM6-specific siRNA on adenovirus infection in vivo, we employed a live animal imaging system (IVIS 100; Xenogen, Caliper Life Sciences). Pretreatment with CEACAM6-specific siRNA, compared with control siRNA or PBS, enhanced reporter gene expression (P < 0.001) from an adenovirus vector at 24 hours about 3-fold after 1 intratumoral injection of replicating adenovirus vector (Figure 6B). However, the reporter gene expression in the CEACAM6-siRNA–pretreated group was just slightly higher than that in the other 2 groups at 48 and 72 hours and declined to a level similar to that in controls thereafter. This suggests that the increased adenovirus infection at 24 hours in CEACAM6-siRNA–treated tumors did not induce more infectious virus to infect neighboring cells because of restoration of CEACAM6 expression. There might be other host factors affecting the penetration of oncolytic adenovirus in vivo. To extend this result to an efficacy study, we established CEACAM6-overexpressing xenografts of Suit-2 cancer cells in nude mice and then treated them with siRNA (systemically) or/and oncolytic adenovirus (intratumorally). As shown in Figure 6C, treatment with oncolytic adenovirus alone or control siRNA had no therapeutic effect when compared with the administration of PBS. CEACAM6-siRNA and the combination of control siRNA and oncolytic adenovirus extended the tumor progression–free time, but the difference did not reach statistical significance compared with the control PBS group (P = 0.20). Of all the treatments, only CEACAM6-siRNA plus Ad5 led to significantly prolonged survival, measured as time to tumor progression (P < 0.05).
Figure 6. Systemic delivery of CEACAM6-siRNA–enhanced adenovirus infection; and antitumor efficacy in vivo.
(A) CEACAM6 expression in Suit-2 xenografts after i.p. injection of PBS, control siRNA, and CEACAM6-specific SMARTpool siRNA by immunohistochemical staining; original magnification, ×200. CEACAM6 expression was suppressed by 37% at 24 hours after systemic delivery of CEACAM6-specific siRNA. There was no significant reduction of CEACAM6 expression in tumors treated with PBS and control siRNA over time. (B) Viral replication of adenovirus in vivo as assessed by live animal imaging. The average radiance of the fluorescent signal from each experimental group was plotted for 5 days after intratumoral injection of replicating adenovirus expressing red fluorescent protein (RFP). p/s/cm2/sr, photons/second/cm2/steradian. (C) Prolonged survival induced by CEACAM6-specific siRNA and oncolytic adenovirus. The systemic pretreatment with CEACAM6-specific siRNA plus oncolytic adenovirus induced a statistically significant survival time (P < 0.05) compared with PBS, oncolytic virus, CEACAM6-siRNA, or control siRNA alone or combination of control siRNA and oncolytic adenovirus. The percentage of mice free from tumor progression (tumor volume <1,000 μl) at each time point was estimated using the Kaplan-Meier method; 6 animals per group.
Discussion
CEACAM6 is a glycosylphosphatidylinositol-linked (GPI-linked) immunoglobulin superfamily member, found principally on neutrophils and some epithelia (26). There is accumulating evidence that CEACAM6 has functional importance in tumorigenesis, including disrupting tissue architecture and affecting cell polarity, differentiation, anti-anoikis activity, tumor invasion, metastasis, and even chemoresistance (17, 19, 20, 22, 27, 28). Inhibition of CEACAM6 expression by RNA interference or antibody targeting can inhibit tumor adhesion, growth, invasion, and metastasis, resulting in improved survival of mice with metastases (18, 29, 30). It was recently reported that CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting the hypothesis that E. coli colonizes the ileal mucosa in Crohn disease (31). However, little is known about the interaction of CEACAM6 and adenovirus infection. In the present study, we demonstrate, for the first time to our knowledge, that CEACAM6 affects adenovirus infectivity by blocking adenovirus trafficking via the Src pathway. These findings have important implications for the development of new biomarkers to predict the response of cancer patients to adenovirus-based therapy and new therapeutics to improve the potency of adenovirus-based gene therapy. It is becoming increasingly evident that the expression of host genes determines the efficiency of adenovirus infection and the outcome of adenovirus-based therapeutics. In this study, we found that a common tumor-associated gene, CEACAM6, functionally affected adenovirus infection (Figure 3). Our stepwise dissection of the adenovirus life cycle in CEACAM6-modulated cells demonstrated that CEACAM6 does not affect adenovirus receptor expression, attachment, and internalization, but rather affects adenovirus trafficking.
Intact microtubules play a very important role in adenovirus trafficking, since elimination of cytoplasmic dynein microtubule function prevents adenovirus translocation to the nucleus and eliminates the characteristic saltatory dynamic motility of the labeled capsid (24, 32–35). In the current study, it was found that overexpression of CEACAM6 can alter the distribution and reduce the expression of cytoskeletal proteins such as tubulin and dynactin, resulting in blockage of adenovirus cytoplasmic trafficking to the nucleus (Figures 4 and 5). Surprisingly, we found that overexpression of CEACAM6 downregulated Src activity and that the Src-selective inhibitor PP2 can disturb the cytoskeleton and interfere with adenovirus infection. This is contrary to a previous report that CEACAM6 cross-linking increased Src activity (28). It is known that several GPI-anchored proteins, including CEACAM family members, have no transmembrane or intracellular domains, but they are still able to modulate intracellular signaling events and to influence cell activities. The molecular mechanisms by which CEACAM6 affects Src are yet to be determined. Caveolin-1 acts as an adaptor between GPI and c-Src signaling pathways (36). It was reported that modulation of c-Src tyrosine kinase activity by CEACAM6 is an acaveolin-1–dependent process (28), and we found that overexpression of CEACAM6 increased the phosphorylation of caveolin-1 and led to more Csk translocation into the cell membrane of cancer cells overexpressing CEACAM6 (R. Gangeswaran, unpublished observations), consistent with a previous report (37). These observations imply a possible mechanism by which CEACAM6 interacts with caveolin-1 and Csk and downregulates Src activity. Given that CEACAM6 may interact with multiple signaling pathways that might also have implications in adenovirus infection, further studies are needed to dissect these networks. In addition, whether CEACAM6 affects adenovirus early gene expression and DNA replication after the delivery of the viral DNA core into the nucleus also requires further investigation to determine more effective approaches to enhance the oncolysis of adenovirus and expand our knowledge of the interaction of host cells and adenovirus.
While adenoviral infection normally results in mild illness in humans, more severe effects are seen in immunocompromised patients. Insights into host factors that affect the adenovirus life cycle may result in development of new antiviral drugs directed against cellular, rather than viral, targets. In the present study, we found that an Src-selective inhibitor dramatically reduced adenovirus replication in normal epithelial cells, suggesting that Src-specific inhibitors may be effective against adenovirus infections.
RNA interference silences genes with a high degree of specificity and may provide a basis for molecularly targeted anticancer therapy (38). Systemic administration of CEACAM6-specific siRNA induced suppression of tumor growth, inhibition of metastasis, and improved survival in a human pancreatic cancer animal model (29) but did not show significant antitumor efficacy in the Suit-2 tumor model in our study. In the study by Duxbury et al. (29), mice were treated with siRNA twice weekly for 6 weeks. In our experiment, we treated the animals with siRNAs for a total of 5 times within 1 week (3 times before the first injection of oncolytic adenovirus, then 1 injection each before the second and third administration of oncolytic adenovirus) because we wished primarily to investigate whether downregulation of CEACAM6 expression could enhance the antitumor potency of oncolytic adenovirus, based on the proposed mechanism of effects on adenovirus trafficking. The combination of CEACAM6-specific siRNA and oncolytic adenovirus significantly prolonged the survival of mice with Suit-2 xenografts when compared with monotherapy, despite a low efficiency of siRNA delivery, with just 40% suppression of CEACAM6 expression in vivo. Other delivery methods, such as combining siRNA with polymers or lipidoids, may improve delivery (39). CEACAM6 inhibition has the potential to improve adenovirus-based gene therapy.
Taken together, the results of the present study have identified CEACAM6 as a new tumor-associated gene that affects the infectability of cancer cells to adenovirus vector. There are important implications for CEACAM6 and CEACAM6-associated signaling pathways as potential targets for the development of new biomarkers to predict the response of patients to adenovirus-based gene therapy and new therapeutics to enhance the efficacy of adenovirus-based gene therapy.
Methods
Cell lines and cell culture.
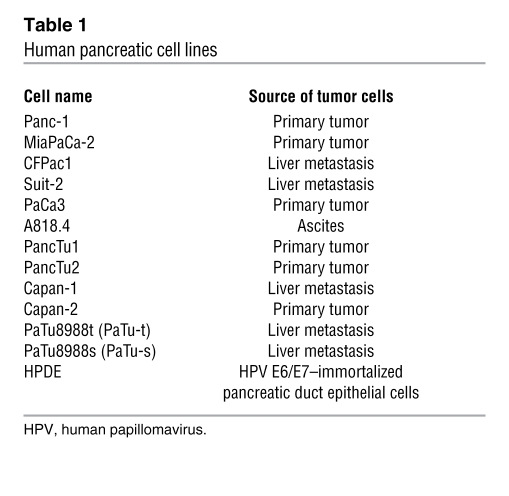
The pancreatic cell lines used are listed in Table 1. The human lung cancer cell line A549 and HEK cells (a subclone named JH-293 was used in the study) were obtained from the Cancer Research UK Central Cell Service (Clare Hall). All cell lines were grown at 37°C with 5% CO2. All cells except Capan-2 and HPDE were maintained in DMEM supplemented with 10% FBS, 50 μg/ml streptomycin, and 50 μg/ml penicillin. Capan-2 cells were maintained in McCoy’s modified medium (ATCC) with 10% FBS, and HPDE cells were maintained in keratinocyte serum–free medium (Gibco, Invitrogen) supplemented with 5 ng/ml EGF and 0.1 mg/ml bovine pituitary extract.
Table 1 .
Human pancreatic cell lines
Establishment of CEACAM6-expressing stable cell lines.
PaTu8988t and HCT116 cells were seeded at 1 × 106 per 10-cm-diameter plate and transfected using FuGENE6 (Roche Diagnostics) in a 3:1 ratio with PML1-Hyg or PML1-CEACAM6 (provided by Clifford Stanners, McGill University, Montreal, Quebec, Canada) according to the manufacturer’s protocol. The stable cell lines were established following selection with 200 μg/ml Hygromycin B (Invitrogen). The expression of CEACAM6 was induced with 200 μg/ml zinc sulfate (Sigma-Aldrich).
Affymetrix analysis.
RNA was isolated from PaTu8988t and PaTu8988s cells using TRIzol solution (Gibco, Invitrogen) following the manufacturer’s recommendations. The Affymetrix analysis was performed as previously described (40) by using U133 Plus 2.0 GeneChips.
Viruses.
Serotype 5 human wild-type adenovirus Ad5, the replication-defective Ad5 mutant dl312, and Ad-CMV-GFP nonreplicating adenoviruses were stored, purified, and titrated in our laboratory as previously described (41, 42). Replicating adenovirus expressing red fluorescent protein was provided by Gunnel Hallden and Daniel Oberg (Institute of Cancer, Barts and The London School of Medicine and Dentistry). For all experiments, except for in vivo imaging, the oncolytic adenovirus indicated is serotype 5 human adenovirus.
Viral infection and replication assay.
Cells were seeded in 6-well plates (1× 105 to 3 × 105 cells/well according to the growth properties of each cell line) and infected with wild-type adenovirus Ad5. Samples were harvested in triplicate at 24-hour intervals up to 144 hours. Viral replication was detected by TCID50 (50% tissue culture infective dose) as described previously (41).
In vitro cell survival assay.
For cell survival assays, 1 × 103 to 5 × 103 cells were seeded in each well of 96-well plates in 90 μl of medium and 16 hours later infected with Ad5 at a serial dilution. Cell survival was determined by MTS assay (Promega) 6 days after infection, and the EC50 values of wild-type adenovirus in different tumor cell lines were obtained as previously described (41). All assays were performed at least 3 times, and in each individual assay each concentration of the virus was tested in sextuplicate.
Western blot analysis.
Immunoblotting was performed on the following pancreatic cell lines: PaTu8988t, HPDE, Panc1, MiaPaCa-2, Suit-2, PaTu8988s, Capan-1, and Capan-2. Total protein lysate (15 μg) was size-fractionated on 4%–12% NuPage gradient Bis-Tris Gel (Invitrogen) and blotted onto a nitrocellulose membrane (Millipore) for 45 minutes at 20 V. For immunodetection, the blots were incubated for 45–60 minutes at room temperature with 1:1,000 dilution of mouse monoclonal CEACAM6 antibody (Abcam) followed by 1 hour incubation with HRP-conjugated secondary antibody (Autogen Bioclear). c-Src expression was analyzed using 30 μg total protein with 1:500 dilutions of rabbit anti–Src-529 or rabbit anti–Src-418 or mouse anti–pan-Src (Abcam), followed by 1 hour incubation with appropriate HRP-conjugated secondary antibodies (Autogen Bioclear). Detection of the bound antibody was carried out using ECL Western blotting detection reagents (Amersham Biosciences, GE Healthcare). Anti-actin antibody was purchased from Santa Cruz Biotechnology Inc.
Analysis of CAR expression by FACS.
Cells (1 × 106) of each cell line (including the positive control A549 cell line) were harvested and washed with cold PBS, then blocked with rabbit serum for 30 minutes, followed by incubation for 1 hour on ice with monoclonal antibody Rcmb for CAR (ATCC). Cells were washed with PBS and incubated with a secondary FITC-conjugated rabbit anti-mouse antibody (Dako), then washed and resuspended with 50 μg/ml propidium iodide (PI) in FACS buffer (PBS, 2% FCS, 1 mM EDTA). Cells were then processed through a FACS scanner, and the percentage difference of the mean of the FITC fluorescence intensity between the sample and the matched control was obtained after 20,000 events were scored.
Assessment of infectivity of adenovirus by FACS.
For infectivity studies, 1 × 105 to 2 × 105 cells were seeded in each well of 6-well plates in the normal medium and incubated at 37°C/10% CO2 overnight. Cells were infected with Ad-CMV-GFP or dl312 adenovirus at a MOI of 100 pt/cell for 48 hours, followed by trypsinization and resuspension in FACS buffer with 50 μg/ml PI. The percentage of GFP-positive cells was analyzed after 20,000 events were scored from viable cells.
RNA interference.
A 6-well plate was seeded with 1 × 105 Suit-2 cells/well in antibiotic-free E4 medium supplemented with 5% FCS. After 24 hours, cells were transfected with Custom SMARTpool CEACAM6 siRNA (Dharmacon, Thermo Scientific) by using DharmaFECT siRNA Transfection Reagent 1 according to the manufacturer’s instructions. Experiments were carried out with 50-nM and 100-nM concentrations of siRNA, and the knockdown was monitored by real-time quantitation for mRNA levels and by Western blot analysis for protein levels. For control experiments, siCONTROL Non-Targeting siRNA Pool (Dharmacon, Thermo Scientific) was used under the same conditions.
For virus killing experiments, the siRNA experiments were performed in 24-well plates with 5 × 104 cells/well. After 72 hours of siRNA transfections, cells were treated with Ad5 adenovirus at MOI of 0, 50, 100, and 200 in E4 containing 5% FCS. The cells were incubated for 72 hours, and MTS assays were performed as described above. The control and the CEACAM6 siRNA experiments were carried out in separate wells of the same plate. Three separate experiments were carried out, each in duplicate.
Real-time PCR for quantitation of CEACAM6.
qPCR was carried out using the Applied Biosystems 7500 Real-Time PCR System and the TaqMan 5′ nuclease assay (Applied Biosystems). Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacture’s protocol. The cDNAs were synthesized from 1 μg of total RNA using a MultiScribe kit (Applied Biosystems) with random hexamers. TaqMan reactions were performed in triplicate using TaqMan PCR master mix (Applied Biosystems) with 10 ng/μl cDNA per reactions. Primers and probes were purchased from Applied Biosystems (Hs00366002_m1 for CEACAM6 [80-bp amplicon] and 4319413E for 18S endogenous control). Negative controls were run to ensure that no amplification occurred in the absence of cDNA. CEACAM6 and 18S assays were run in a multiplex reaction. Single and multiplex standard curves were produced with target and endogenous genes to test reaction efficiency of the assay. The standard curve was produced by using a 5-fold serial dilution of universal reference RNA (Stratagene). Data were analyzed using Sequence Detection Software version 1.3 (Applied Biosystems). The relative expression was normalized to A549 values.
TaqMan qPCR assay for Ad5 binding, internalization, and trafficking to the nuclei.
All steps were carried out with PBS buffer containing 1% BSA. Appropriate cells were suspended (2 × 105 in 100 μl) in test tubes and incubated with 100 μl of buffer alone or buffer containing Ad5 (5,000 virus particles/cell) at 4°C with vigorous shaking for 1 hour. For the binding assay, the cells were washed 3 times with cold buffer to remove unbound virus particles and collected for total DNA preparation using the QIAamp DNA Blood Mini Kit (QIAGEN). For internalization assays, the washed cells were placed at 37°C for various intervals, the attached but uninternalized viral particles were removed with 2 mg/ml subtisilin (Sigma-Aldrich), and the samples were collected for DNA preparation. Nuclear fractions were separated using the NE-PER nuclear and cytoplasmic kit from Pierce (Thermo Scientific). The viral genome copy number was determined using TaqMan quantitative real-time PCR with E1A primers, as described below. Quantitative PCR was carried out using the Applied Biosystems 7500 Real-Time PCR system and the TaqMan 5′ nuclease assay (Applied Biosystems). E1A TaqMan reactions were performed in triplicate using TaqMan PCR Master Mix (Applied Biosystems) with 10 ng DNA per reaction. Primers were purchased from Sigma-Aldrich (E1A, 5′-TGTACCGGAGGTGATCGATCT-3′; E1A, 3′-TCGTCACTGGGTGGAAAGC-5′) and the probe from Applied Biosystems (E1A probe, VIC-ACCTGCCACGAGGC-MGB) to give a 57-bp amplicon. Negative controls were run to ensure that no amplification occurred in the absence of DNA. Multiple standard curves were produced with target (Ad5 pure viral DNA) to test reaction efficiency and reproducibility of the assay. An absolute standard curve with a dynamic range of 5 × 106 to 5 × 102 E1A copies was produced. Data were analyzed using Sequence Detection Software version 1.3 (Applied Biosystems). All experiments were repeated at least 3 times.
TEM analysis.
Appropriate cells were suspended in PBS buffer containing 1% BSA (1 × 106 in 100 μl) in test tubes and incubated with 100 μl of buffer alone or buffer containing Ad5 (10,000 virus particles/cell) at 4°C with vigorous shaking for 1 hour. The cells were washed 3 times with cold buffer to remove unbound virus particle and placed at 37°C for various time intervals, then washed 3 times with phosphate buffer and fixed in phosphate-buffered 4% glutaraldehyde for a minimum of 2 hours. The cells were pelleted at 420 g for 5 minutes, and the pellets were mixed with 2% agarose at 80°C and allowed to cool to room temperature. One-millimeter cubes were excised from agarose and placed in phosphate buffer overnight. After primary fixation in glutaraldehyde, specimens were postfixed in osmium tetroxide and dehydrated in a graded series of industrial methylated spirits (IMS) alcohol. The IMS alcohol was then cleared with propylene oxide and tissue blocks gradually infiltrated with Araldite. Ultrathin sections were post-stained with uranyl acetate and lead citrate and imaged with a JEOL JEM 1230 electron microscope fitted with a Soft Imaging Solutions Morada digital camera (Olympus).
PP2 treatment.
For PP2 (obtained from Merck Chemicals Ltd.) treatment, 5 × 104 cells/well were plated into 24-well plates. After 24 hours, cells were exposed to 10 μM PP2, which has been shown previously to potently inhibit c-Src activity in BxPC3 cells (19, 20), for 2 hours prior to performing further assays. DMSO served as a vehicle control.
In vitro c-Src tyrosine kinase assay.
c-Src tyrosine kinase activity was determined in triplicate using a commercially available kinase assay kit (Sigma-Aldrich), according to the manufacturer’s instructions. c-Src immunoprecipitates (20 μg total protein) were prepared from cell lysates using anti–c-Src monoclonal antibody (Autogen Bioclear) immobilized onto protein G–Sepharose beads (Invitrogen). Immunoprecipitates were washed and dissolved in tyrosine kinase buffer (final solution containing 0.3 mM ATP) and incubated for 30 minutes in 96-well plates coated with tyrosine kinase substrate solution (poly-Glu-Tyr). Phosphorylated substrate was quantified by chromogenic detection using HRP-conjugated anti-phosphotyrosine antibody. Optical densities were determined at 492 nm using an Opsys MR (Dynex Technologies Ltd.). c-Src kinase activity was compared with an EGFR standard.
Conjugation of fluorescent dye to adenovirus particles.
The Alexa Fluor 555 carboxylic acid, succinimidyl ester (Invitrogen), was conjugated to our Ad5 stock using a protocol identical to that described previously (43), except that we labeled wild-type adenovirus instead of nonreplicating adenovirus vector.
Infection of cells with fluorophore-labeled Ad5.
Monolayers of stable cell lines PaTu8988t-Hyg and PaTu8988t-CEACAM6 were grown in chamber slides (10,000 cells/well) with E4 and the supplements as mentioned above. The next day, the slides were incubated at 4°C for an hour before infection with labeled Ad5, and the medium was replaced with fresh infection medium (E4 plus labeled Ad5 at a concentration of 1,000 pt/ml) and incubated at 4°C for an hour to allow the virus to bind to the cells. After an hour of cold incubation, the cells were washed twice with cold PBS, which was then replaced with warm medium (full medium containing E4 and the supplements), and the slides were incubated at 37°C for various time intervals, washed twice with prewarmed PBS, and immediately fixed in methanol at –20°C for 10 minutes. The immunostaining was performed as described above. Alpha-tubulin (Sigma-Aldrich) antibody was used at 1:500 dilution. The confocal analysis was carried out as described above.
Immunofluorescence.
The cells were grown overnight on chamber slides (Lab-Tek; Nalge Nunc International) with 5% FCS (Life Technologies). After 2 washes with PBS, cells were fixed in methanol for 15 minutes at room temperature. To visualize F-actin, we fixed the cells in 4% formaldehyde. Cells were then washed with PBS. Nonspecific binding sites were blocked with 1% BSA in PBS for 30 minutes at room temperature. Subsequently, cells were incubated with a 1:500 dilution of mouse anti-CEACAM6 antibody (Abcam), with a 1:500 dilution of rabbit anti-tubulin (Sigma-Aldrich), or a 1:500 dilution of goat anti–dynactin p62 (Autogen Bioclear) for 1 hour at room temperature. After 3 washes in PBS, the primary antibodies were detected with Alexa Fluor 488–conjugated donkey anti-mouse, Alexa Fluor 546–conjugated goat anti-rabbit, or Alexa Fluor 546–conjugated donkey anti-goat secondary antibody, respectively (Molecular Probes, Invitrogen) at a dilution of 1:1,000. DNA was stained with DAPI (Molecular Probes, Invitrogen) at a dilution of 20 μg/ml in PBS. To visualize F-actin, we used 50 μg/ml phalloidin-TRITC (Sigma-Aldrich). After 3 washes, slides were mounted using Permafluor aqueous mounting medium (Immunotech). All experiments were carried out in the absence of primary, secondary, or both antibodies as negative controls. A series of optical sections were taken with a Zeiss LSM 510 confocal microscope and projected to single images using LSM 510 software (Zeiss). Identical acquisition methods were used for all samples to allow direct comparison of the resulting images. Photomontages were created using Adobe Photoshop 5.1 software.
In vivo effect of siRNA on CEACAM6 expression in Suit-2 xenograft tumor model.
All animal studies were approved by the Home Office of the UK and the Animal Welfare and Research Ethics Committee of Zhengzhou University. For determination of the efficiency of CEACAM6-specific siRNA on the downregulation of CEACAM6 in vivo, tumors were grown in one flank of 4- to 6-week-old BALB/c nu/nu mice (female) by subcutaneous implantation of 1 × 106 Suit-2 cells before CEACAM6 siRNA or control siRNA (150 ng/g/injection in 100-μl solution of PBS) was injected i.p. daily for 3 days. The control groups were treated with PBS only. Treatments were initiated when tumors were at least 100 μl in volume; the animals were sacrificed at 24, 48, and 72 hours after treatment (each group/time point comprised 3 mice) and tumors harvested and frozen in isopentane cooled in liquid nitrogen before immunohistochemistry was performed for CEACAM6 expression.
Immunohistochemical analysis.
Immunohistochemistry was performed on 4-μm-thick frozen sections of tissues with 1:200 dilution of CEACAM6 antibody (1 mg/ml; Abcam). After 1 hour incubation with primary antibody, linker antibody (rabbit monoclonal, anti-mouse IgG1 and IgG2 Fc-specific, from Epitomics) was used as the secondary antibody. The DAB Detection Kit was used according to the protocols provided for the Ventana Moleclular Discovery System. CEACAM6 expression was calculated as the percentage of tumor cells showing downregulation of CEACAM6 in the largest cross-section of tumor xenografts (intensity was significantly reduced compared with that in nontreated Suit-2 xenografts).
Evaluation of viral replication and antitumor efficacy of adenovirus in vivo following treatment with CEACAM6-specific siRNA and control siRNA.
Suit-2 cells (1 × 106) were implanted subcutaneously into the right flank of 4- to 5-week-old BALB/c nude female mice (n = 36). When tumors reached about 100 μl, mice were regrouped by tumor size at the time of treatment initiation in all cases (t test for tumor volumes, P > 0.8). Treatments with PBS, control siRNA, and CEACAM6 siRNA were administered on day 1, 2, and 3 by i.p. infection as described previously (25). For assessment of viral replication in vivo, 1 × 1010 pt of replicating adenovirus vector expressing red fluorescent protein in 50 μl was injected intratumorally on day 4 after each injection of siRNAs and PBS. Viral replication in tumor tissues was monitored by a live animal imaging system (IVIS 100; Xenogen, Caliper Life Sciences) for fluorescent signal, on days 1, 2, 3, 4, and 5 after viral treatment. For the efficacy study, the PBS, control siRNA, and CEACAM6 siRNA treatments were administered on days 1, 2, 3, 5, and 7 by i.p. injection, and 1 × 1010 pt of oncolytic adenovirus Ad5 was injected intratumorally on days 4, 6, and 8. Tumor volumes were estimated [volume = (width × length2 × π)/6] twice weekly until mice were sacrificed when tumor volume reached 1.44 cm3 or symptomatic tumor ulceration occurred (according to animal use regulations of the UK Home Office and the Department of Science and Technology of Henan Province, People’s Republic of China). Survival analysis expressed as time to progression (tumor volume ≥1,000 μl) was performed according to the Kaplan-Meier method (log-rank test for statistical significance).
Statistics.
All the statistical comparisons of adenovirus receptor expression, infectivity, cytotoxicity, and replication between cancer cells were carried out by unpaired 2-tailed Student’s t test. In xenograft tumor studies, assessment of viral replication was analyzed by 2-way ANOVA with Bonferroni post-tests, and P values of less than 0.05 were considered to be statistically significant for all tests. All data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
This project is supported by Cancer Research UK (C633-A6253/A6251 programme grant) and the Nature Sciences Foundation of China (no. 30530800). We appreciate the generous gift of valuable materials from Clifford Stanners of McGill University. Many thanks go to Katherine Excoffon of the University of Iowa for her useful protocol of TEM and Graham McPhail of Queen Mary, University of London in Whitechapel for help with TEM operation. We are very grateful to Gunnel Hallden, Kairbaan Hodivala-Dilke, Iain McNeish, and Ian Hart for their critical comments. Finally, we thank Keyur Trivedi and Mohammed Ikram for their excellent work in immunohistochemistry.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: CAR, Coxsackie and adenovirus receptor; CEACAM6, carcinoembryonic antigen–related cell adhesion molecule 6; GPI, glycosylphosphatidylinositol; pt, particle(s); qPCR, quantitative real-time PCR; TEM, transmission electron microscopy.
Citation for this article: J. Clin. Invest. 119:1604–1615 (2009). doi:10.1172/JCI37905
References
- 1.Garber K. China approves world’s first oncolytic virus therapy for cancer treatment. J. Natl. Cancer Inst. 2006;98:298–300. doi: 10.1093/jnci/djj111. [DOI] [PubMed] [Google Scholar]
- 2.Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Ther. 2001;8:89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- 3.Kim S., Peng Z., Kaneda Y. Current status of gene therapy in Asia. Mol. Ther. 2008;16:237–243. doi: 10.1038/sj.mt.6300336. [DOI] [PubMed] [Google Scholar]
- 4.Rein D.T., Breidenbach M., Curiel D.T. Current developments in adenovirus-based cancer gene therapy. Future Oncol. 2006;2:137–143. doi: 10.2217/14796694.2.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parato K.A., Senger D., Forsyth P.A., Bell J.C. Recent progress in the battle between oncolytic viruses and tumours. Nat. Rev. Cancer. 2005;5:965–976. doi: 10.1038/nrc1750. [DOI] [PubMed] [Google Scholar]
- 6.Hermiston T. A demand for next-generation oncolytic adenoviruses. Curr. Opin. Mol. Ther. 2006;8:322–330. [PubMed] [Google Scholar]
- 7.Mulvihill S., et al. Safety and feasibility of injection with an E1B-55 kDa gene-deleted, replication-selective adenovirus (ONYX-015) into primary carcinomas of the pancreas: a phase I trial. Gene Ther. 2001;8:308–315. doi: 10.1038/sj.gt.3301398. [DOI] [PubMed] [Google Scholar]
- 8.Kirn D. Replication-selective oncolytic adenoviruses: virotherapy aimed at genetic targets in cancer. Oncogene. 2000;19:6660–6669. doi: 10.1038/sj.onc.1204094. [DOI] [PubMed] [Google Scholar]
- 9.Hecht J.R., et al. A phase I/II trial of intratumoral endoscopic ultrasound injection of ONYX-015 with intravenous gemcitabine in unresectable pancreatic carcinoma. Clin. Cancer Res. 2003;9:555–561. [PubMed] [Google Scholar]
- 10.Li E., Stupack D., Klemke R., Cheresh D., Nemerow G. Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J. Virol. 1998;72:2055–2061. doi: 10.1128/jvi.72.3.2055-2061.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li E., Stupack D., Bokoch G., Nemerow G. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 1998;72:8806–8812. doi: 10.1128/jvi.72.11.8806-8812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suomalainen M., Nakano M., Boucke K., Keller S., Greber U. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001;20:1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumann M., Dobbelstein M. Adenovirus-induced extracellular signal-regulated kinase phosphorylation during the late phase of infection enhances viral protein levels and virus progeny. Cancer Res. 2006;66:1282–1288. doi: 10.1158/0008-5472.CAN-05-1484. [DOI] [PubMed] [Google Scholar]
- 14.Kim J., et al. Antitumoral effects of recombinant adenovirus YKL-1001, conditionally replicating in alpha-fetoprotein-producing human liver cancer cells. Cancer Lett. 2002;180:23–32. doi: 10.1016/S0304-3835(02)00017-4. [DOI] [PubMed] [Google Scholar]
- 15.Xia H., Anderson B., Mao Q., Davidson B.L. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. . J. Virol. 2000;74:11359–11366. doi: 10.1128/JVI.74.23.11359-11366.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier O., et al. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilantzis C., DeMarte L., Screaton R.A., Stanners C.P. Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia. 2002;4:151–163. doi: 10.1038/sj.neo.7900201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duxbury M.S., Ito H., Zinner M.J., Ashley S.W., Whang E.E. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23:465–473. doi: 10.1038/sj.onc.1207036. [DOI] [PubMed] [Google Scholar]
- 19.Duxbury M.S., et al. Overexpression of CEACAM6 promotes insulin-like growth factor I-induced pancreatic adenocarcinoma cellular invasiveness. Oncogene. 2004;23:5834–5842. doi: 10.1038/sj.onc.1207775. [DOI] [PubMed] [Google Scholar]
- 20.Duxbury M.S., et al. A novel role for carcinoembryonic antigen-related cell adhesion molecule 6 as a determinant of gemcitabine chemoresistance in pancreatic adenocarcinoma cells. Cancer Res. 2004;64:3987–3993. doi: 10.1158/0008-5472.CAN-04-0424. [DOI] [PubMed] [Google Scholar]
- 21.Le L.P., et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J. Natl. Cancer Inst. 2006;98:203–214. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 22.Duxbury M.S., Ito H., Ashley S.W., Whang E.E. c-Src-dependent cross-talk between CEACAM6 and alphavbeta3 integrin enhances pancreatic adenocarcinoma cell adhesion to extracellular matrix components. Biochem. Biophys. Res. Commun. 2004;317:133–141. doi: 10.1016/j.bbrc.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Greber U.F., Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Suomalainen M., et al. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Compagno D., et al. SIRNA-directed in vivo silencing of androgen receptor inhibits the growth of castration-resistant prostate carcinomas. PLoS ONE. 2007;2:e1006. doi: 10.1371/journal.pone.0001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholzel S., et al. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am. J. Pathol. 2000;156:595–605. doi: 10.1016/S0002-9440(10)64764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ordonez C., Screaton R.A., Ilantzis C., Stanners C.P. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 2000;60:3419–3424. [PubMed] [Google Scholar]
- 28.Duxbury M.S., Ito H., Ashley S.W., Whang E.E. CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J. Biol. Chem. 2004;279:23176–23182. doi: 10.1074/jbc.M402051200. [DOI] [PubMed] [Google Scholar]
- 29.Duxbury M.S., et al. 2004Systemic siRNA–mediated gene silencing: a new approach to targeted therapy of cancer. Ann. Surg. 240667–674; discussion 675–676. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blumenthal R.D., Hansen H.J., Goldenberg D.M. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen). Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
- 31.Barnich N., et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. . J. Clin. Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miles B.D., Luftig R.B., Weatherbee J.A., Weihing R.R., Weber J. Quantitation of the interaction between adenovirus types 2 and 5 and microtubules inside infected cells. Virology. 1980;105:265–269. doi: 10.1016/0042-6822(80)90177-4. [DOI] [PubMed] [Google Scholar]
- 33.Mabit H., et al. Intact microtubules support adenovirus and herpes simplex virus infections. . J. Virol. 2002;76:9962–9971. doi: 10.1128/JVI.76.19.9962-9971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leopold P.L., et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 35.Kelkar S.A., Pfister K.K., Crystal R.G., Leopold P.L. Cytoplasmic dynein mediates adenovirus binding to microtubules. J. Virol. 2004;78:10122–10132. doi: 10.1128/JVI.78.18.10122-10132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargiacomo M., Sudol M., Tang Z., Lisanti M.P. Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. . J. Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao H., Courchesne W.E., Mastick C.C. A phosphotyrosine-dependent protein interaction screen reveals a role for phosphorylation of caveolin-1 on tyrosine 14: recruitment of C-terminal Src kinase. J. Biol. Chem. 2002;277:8771–8774. doi: 10.1074/jbc.C100661200. [DOI] [PubMed] [Google Scholar]
- 38.Lemoine N.R. Silencing RNA: a novel treatment for pancreatic cancer? Gut. 2005;54:1215–1216. doi: 10.1136/gut.2005.067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akinc A., et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capurso G., et al. Gene expression profiles of progressive pancreatic endocrine tumours and their liver metastases reveal potential novel markers and therapeutic targets. Endocr. Relat. Cancer. 2006;13:541–558. doi: 10.1677/erc.1.01153. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., et al. E3 gene manipulations affect oncolytic adenovirus activity in immunocompetent tumor models. Nat. Biotechnol. 2003;21:1328–1335. doi: 10.1038/nbt887. [DOI] [PubMed] [Google Scholar]
- 42.Hallden G., et al. Novel immunocompetent murine tumor models for the assessment of replication-competent oncolytic adenovirus efficacy. Mol. Ther. 2003;8:412–424. doi: 10.1016/S1525-0016(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 43.Leopold P.L., et al. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.