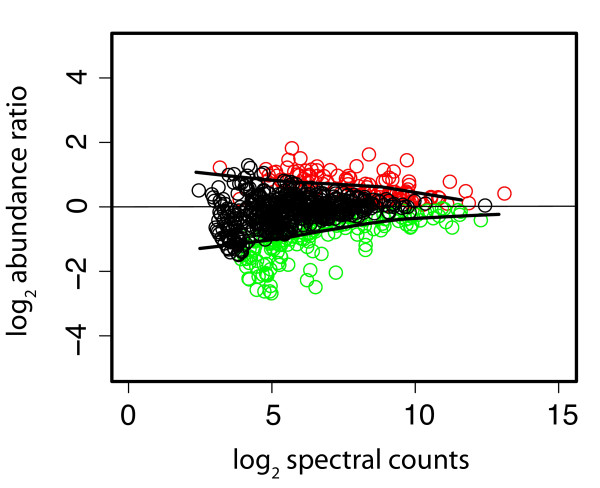
Figure 2.
Pseudo M versus A plot [28,29] of the average protein abundance ratios over all replicates for the P. gingivalis-F. nucleatum-S. gordonii/P. gingivalis comparison versus total abundance as estimated by spectral counting. Color codes: red, P. gingivalis protein is over-expressed in the P. gingivalis-F. nucleatum-S. gordonii community relative to P. gingivalis alone; green, P. gingivalis protein is under-expressed in the community relative to P. gingivalis alone; black, no significant abundance change. Solid black lines represent a LOWESS curve fit [30] to the biological replicates of P. gingivalis alone, and represent the upper and lower boundaries of the experimentally observed error regions or null distributions associated with the relative abundance ratio calculations. Proteins coded as either red or green were determined to be significantly changed at the q-value [24] cut-off value of 0.01. Thus, the G-test predictions [56] were in good agreement with the curve fitting procedure. Details regarding hypothesis testing procedures can be found in Methods and in the explanatory notes to the data tables [see Additional File 1].